Abstract 抽象
Nutrients and energy have emerged as central modulators of developmental programmes in plants and animals1,2,3. The evolutionarily conserved target of rapamycin (TOR) kinase is a master integrator of nutrient and energy signalling that controls growth. Despite its key regulatory roles in translation, proliferation, metabolism and autophagy2,3,4,5, little is known about how TOR shapes developmental transitions and differentiation. Here we show that glucose-activated TOR kinase controls genome-wide histone H3 trimethylation at K27 (H3K27me3) in Arabidopsis thaliana, which regulates cell fate and development6,7,8,9,10. We identify FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), an indispensable component of Polycomb repressive complex 2 (PRC2), which catalyses H3K27me3 (refs. 6,7,8,10,11,12), as a TOR target. Direct phosphorylation by TOR promotes the dynamic translocation of FIE from the cytoplasm to the nucleus. Mutation of the phosphorylation site on FIE abrogates the global H3K27me3 landscape, reprogrammes the transcriptome and disrupts organogenesis in plants. Moreover, glucose–TOR–FIE–PRC2 signalling modulates vernalization-induced floral transition. We propose that this signalling axis serves as a nutritional checkpoint leading to epigenetic silencing of key transcription factor genes that specify stem cell destiny in shoot and root meristems and control leaf, flower and silique patterning, branching and vegetative-to-reproduction transition. Our findings reveal a fundamental mechanism of nutrient signalling in direct epigenome reprogramming, with broad relevance for the developmental control of multicellular organisms.
营养物质和能量已成为植物和动物发育计划的核心调节剂 1,2,3。雷帕霉素 (TOR) 激酶的进化保守靶标是控制生长的营养和能量信号的主要整合者。尽管 TOR 在翻译、增殖、代谢和自噬中起关键调节作用 2,3,4,5,但人们对 TOR 如何塑造发育转变和分化知之甚少。在这里,我们表明葡萄糖激活的 TOR 激酶控制拟南芥 K27 (H3K27me3) 的全基因组组蛋白 H3 三甲基化,从而调节细胞命运和发育 6,7,8,9,10。我们确定了不依赖受精的胚乳 (FIE),它是多梳抑制复合物 2 (PRC2) 不可或缺的组成部分,它催化 H3K27me3 (参考文献。6,7,8,10,11,12) 作为 TOR 目标。TOR 的直接磷酸化促进 FIE 从细胞质到细胞核的动态易位。FIE 上磷酸化位点的突变消除了全球 H3K27me3 景观,重新编程转录组并破坏了植物的器官发生。此外,葡萄糖-TOR-FIE-PRC2 信号调节春化诱导的花转变。我们提出这个信号轴作为一个营养检查点,导致关键转录因子基因的表观遗传沉默,这些基因指定了芽和根分生组织中的干细胞命运,并控制叶、花和角果模式、分枝和营养到繁殖的转变。 我们的研究结果揭示了直接表观基因组重编程中营养信号传导的基本机制,与多细胞生物的发育控制具有广泛的相关性。
Similar content being viewed by others
其他人正在查看类似内容
Main 主要
Glucose is a universal nutrient and is the main energy supplier, metabolic precursor and source of biomass for most cells. Glucose also functions as an essential regulatory signal that directly or indirectly controls diverse vital processes in multicellular organisms1,2,3,4,5. In photosynthetic plants, CO2 captured from the atmosphere leads to system-wide glucose signalling that has pivotal roles in multiple developmental processes that are vital to agronomically important traits and crop yield, including germination, stem cell to primordial proliferation, organ size and patterning, as well as shoot branching, flowering and fruit and seed development2,3,4,5,13,14,15,16. Notably, plant growth hormones and signalling peptides are ineffective in supporting plant development without the glucose signalling networks2,3,4. However, how glucose signalling modulates cell fates and organogenesis remains largely unknown2,3,4,5,13,14,15,16. The evolutionarily conserved TOR kinase is a major glucose signalling mediator that integrates nutrients and energy, as well as growth factors, hormones and environmental cues to control growth, development and ageing2,3,4,5,17,18,19. Although plant TOR has been implicated in processes such as meristem activation to shoot and root growth and flowering2,3,4,5,15,17,18,19,20,21,22, the molecular mechanisms remain poorly understood.
葡萄糖是一种通用营养素,是大多数细胞的主要能量供应者、代谢前体和生物质来源。葡萄糖还作为一种重要的调节信号发挥作用,直接或间接控制多细胞生物体中的各种重要过程 1,2,3,4,5。在光合作用植物中,从大气中捕获的 CO2 导致全系统葡萄糖信号传导,该信号传导在多个发育过程中起关键作用,这些过程对农艺性状和作物产量至关重要,包括发芽、干细胞到原始增殖、器官大小和模式,以及芽分枝、开花、果实和种子发育2,3,4,5,13,14,15,16。值得注意的是,如果没有葡萄糖信号网络,植物生长激素和信号肽在支持植物发育方面是无效的 2,3,4。然而,葡萄糖信号如何调节细胞命运和器官发生在很大程度上仍然未知 2,3,4,5,13,14,15,16。进化上保守的 TOR 激酶是一种主要的葡萄糖信号传导介质,它整合了营养物质和能量,以及生长因子、激素和环境线索,以控制生长、发育和衰老 2,3,4,5,17,18,19。 尽管植物 TOR 与分生组织激活芽和根生长和开花等过程有关 2,3,4,5,15,17,18,19,20,21,22,但分子机制仍然知之甚少。
Epigenetic regulation on chromatin underlies cell fate specification and developmental transitions, and is a universal mechanism for establishing and maintaining cell and organ identity in plants and animals6,7,8,9,10. PRC2 catalyses histone H3K27me3, triggering epigenetic silencing of key regulatory genes, which are required for cell identity and plasticity to promote differentiation in various developmental programmes6,7,8,10. Although both PRC2 and TOR are vital for diverse plant developmental processes, no direct molecular connection has been established between the PRC2-mediated global H3K27me3 regulation in organogenesis and the TOR-associated dynamic nutrient and energy signalling network.
染色质的表观遗传调控是细胞命运规范和发育转变的基础,是建立和维持植物和动物细胞和器官身份的通用机制 6,7,8,9,10。PRC2 催化组蛋白 H3K27me3,触发关键调节基因的表观遗传沉默,这些基因是细胞身份和可塑性所必需的,以促进各种发育程序中的分化 6,7,8,10。尽管 PRC2 和 TOR 对不同的植物发育过程都至关重要,但 PRC2 介导的器官发生中全球 H3K27me3 调节与 TOR 相关的动态营养和能量信号网络之间尚未建立直接的分子联系。
Glucose–TOR signalling modulates H3K27me3
葡萄糖-TOR 信号调节 H3K27me3
To explore new mechanisms of the complex TOR signalling network in plant development, we conducted unbiased chemical screens for various molecular effects of different levels of TOR kinase deficiency in A. thaliana2,3,4,5. Germinating seeds in liquid nutrient medium were treated with systematically varying concentrations of the potent ATP-competitive TOR inhibitor Torin22,4 at the initiation of postembryonic development. S6K phosphorylation, an evolutionarily conserved indicator of TOR activity4,21, was strongly inhibited by Torin2, starting from 0.1 μM. Higher Torin2 concentrations (5–10 μM) abolished DNA replication, as indicated by quantitative incorporation of the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU)4 and retarded shoot and root development. Notably, decreased TOR activity resulting from treatment with 0.5–10 μM Torin2 gradually decreased levels of the epigenomic mark H3K27me3, which is associated with developmental transitions and organ differentiation6,7,8,9,10. This function of TOR signalling in chromatin regulation was validated genetically in mutant plants with oestradiol-inducible RNA-mediated inhibition of TOR expression4,21 (tor-es), which exhibited suppression of pS6K1(S449), EdU staining, high H3K27me3 and seedling development (Fig. 1a and Extended Data Fig. 1a,b). However, the reduction in H3K27me3 was probably underestimated, partially owing to experimental limitations when DNA replication was abolished by 5–10 μM Torin2 or tor-es4,15 (Fig. 1a).
为了探索植物发育中复杂 TOR 信号网络的新机制,我们对拟南芥中不同水平 TOR 激酶缺陷的各种分子效应进行了无偏的化学筛选 2,3,4,5。在胚胎后发育开始时,用系统变化浓度的有效 ATP 竞争性 TOR 抑制剂 Torin22,4 处理液体营养培养基中发芽的种子。S6K 磷酸化是 TOR 活性 4,21 的进化保守指标,从 0.1 μM 开始被 Torin2 强烈抑制。较高的 Torin2 浓度 (5–10 μM) 消除了 DNA 复制,如胸苷类似物 5-乙炔基-2′-脱氧尿嘧啶 (EdU)4 的定量掺入和芽和根发育迟缓所示。值得注意的是,用 0.5-10 μM Torin2 处理后,TOR 活性降低,表观基因组标记 H3K27me3 的水平逐渐降低,这与发育转变和器官分化有关 6,7,8,9,10。TOR 信号转导在染色质调节中的这种功能在雌二醇诱导的 RNA 介导的 TOR 表达抑制 4,21 (tor-es) 的突变植物中得到了遗传验证,其表现出对 pS6K1(S449) 的抑制、EdU 染色、高 H3K27me3 和幼苗发育的抑制(图 D)。1a 和扩展数据图 1.1a,b)。 然而,H3K27me3 的减少可能被低估了,部分原因是当 DNA 复制被 5-10 μM Torin2 或 tor-es 4,15 消除时,实验限制(图 D)。1a)。
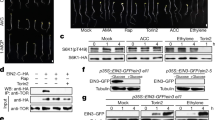
图 1:葡萄糖-TOR 信号传导特异性调节整体 H3K27me3 动力学。
a, Distinct TOR activity thresholds regulate plant development. Seedling development, EdU staining, pS6K1(T449) phosphorylation and H3K27me3 quantification in WT seedlings treated with different concentrations of Torin2 or in tor-es seedlings at 8 days after germination in rich medium. Images are representative of ten seedlings. White scale bar, 2 mm; red scale bar, 25 μm. b, Specific reduction of H3K27me3 levels in 7-day tor-es seedlings (10 μM oestradiol treatment for 3 days). c, Heat map of H3K27me3 or H3K9me2 enrichment in WT and tor-es seedlings. The colour scale indicates reference-adjusted reads per million (RRPM) surrounding peak summit from the ChIP-Rx–seq data. d,e, Glucose–TOR signalling rapidly regulates H3K27me3 dynamics. d, Schematic presentation of experimental design for glucose regulation of TOR activity. h, hour. Scale bar, 2 mm. f,g, TOR but not S6K regulates global H3K27me3 levels. g, S6K activity was monitored by phosphorylation on RPS6 at S237 and S240 (pRPS6(S237) and pRPS6(S240), respectively). Torin2 (T), INK128 (I) and WYE-354 (W) are specific TOR inhibitors; staurosporine (S) and PF-4708671 (P) inhibit S6K1and S6K2 activity. Values show the relative level of histone modifications compared with the corresponding H3 control relative to the intensity in the WT. Experiments were conducted in three biological repeats with similar results. Samples derive from the same experiment and gels and blots were processed in parallel.
a,不同的 TOR 活性阈值调节植物发育。用不同浓度的 Torin2 处理的 WT 幼苗或 tor-es 幼苗在富培养基发芽后 8 天的幼苗发育、EdU 染色、pS6K1(T449) 磷酸化和 H3K27me3 定量。图像代表 10 棵幼苗。白色刻度条,2 毫米;红色刻度尺,25 μm。b,特异性降低 7 天 tor-es 幼苗中的 H3K27me3 水平 (10 μM 雌二醇处理 3 天)。c, WT 和 tor-es 幼苗中 H3K27me3 或 H3K9me2 富集的热图。色标表示 ChIP-Rx-seq 数据中围绕峰顶的参考调整每百万读数 (RRPM)。d,e,葡萄糖-TOR 信号转导快速调节 H3K27me3 动力学。d,TOR 活性葡萄糖调节实验设计的示意图。h, 小时。比例尺,2 mm. f,g,TOR,但不是 S6K 调节整体 H3K27me3 水平。 g,通过 RPS6 在 S237 和 S240 位点的磷酸化监测 S6K 活性 (分别为 pRPS6 (S237) 和 pRPS6 (S240)。Torin2 (T)、INK128 (I) 和 WYE-354 (W) 是特异性 TOR 抑制剂;星形孢菌素 (S) 和 PF-4708671 (P) 抑制 S6K1 和 S6K2 活性。值显示组蛋白修饰与相应的 H3 对照相比相对于 WT 中强度的相对水平。实验在三个生物学重复中进行,结果相似。样品来源于同一实验,凝胶和印迹平行处理。
We next determined the specificity of TOR regulation of chromatin modifications by quantifying major histone methylation marks using immunoblot analyses in seven-day-old wild-type (WT) and tor-es Arabidopsis seedlings. H3K27me3, but not H3K4me3, H3K9me2 or H3K36me3, was specifically reduced in the tor-es mutant (Fig. 1b). We further performed quantitative chromatin immunoprecipitation with an exogenous reference genome followed by deep-sequencing23 (ChIP-Rx–seq). The average density plot and heat map revealed a global reduction of H3K27me3 occupancy in the tor-es mtuant, whereas the H3K9me2 level was unaffected. The reduction of H3K27me3 in the tor-es seedlings was detected throughout the genome (Fig. 1c, Extended Data Fig. 1c,d and Supplementary Table 1), suggesting that the deposition and/or maintenance of H3K27me3 is regulated by TOR.
接下来,我们通过使用免疫印迹分析对 7 日龄野生型 (WT) 和 tor-es拟南芥幼苗中的主要组蛋白甲基化标志进行定量,确定了染色质修饰的 TOR 调节特异性。H3K27me3,而不是 H3K4me3、H3K9me2 或 H3K36me3,在 tor-es 突变体中被特异性还原(图 D)。我们进一步使用外源参考基因组进行定量染色质免疫沉淀,然后进行深度测序23 (ChIP-Rx–seq)。平均密度图和热图显示 H3K27me3 在土壤中的占有率总体降低,而 H3K9me2 水平不受影响。在整个基因组中检测到 tor-es 幼苗中 H3K27me3 的减少(图 D)。1c,扩展数据图1c,d 和补充表 1),表明 H3K27me3 的沉积和/或维持受 TOR 调节。
We then investigated TOR regulation of H3K27me3 in a physiological context by analysing H3K27me3 dynamics in the well-established heterotrophic to photoautotrophic transition stage when the endogenous sugar derived from the seed reserve is depleted three days after germination. Photosynthesis or exogenous glucose could activate TOR, which was monitored by the phosphorylation status of pS6K1(T449) under light4, 15, 21 (Fig. 1d,e). Arabidopsis seedlings germinated and grown in a sugar-free liquid medium naturally entered a reversible quiescent state in growth and development with decreased TOR activity at 3 to 7 days after germination4,15 (Fig. 1d). The H3K27me3 levels were maintained during this period in sugar-containing medium (Extended Data Fig. 1e), but were gradually reduced during the sugar starvation phase, correlating with inactive TOR and stalled seedling development. Glucose addition rapidly stimulated TOR activity and de novo H3K27me3 accumulation within 2 h, suggesting the reprogramming of chromatin states. Consistently, the restoration of H3K27me3 levels was blocked by Torin2 (10 μM) during glucose stimulation (Fig. 1e).
然后,我们通过分析来自种子储备的内源糖在发芽后 3 天耗尽时,在成熟的异养到光合自养过渡阶段的 H3K27me3 动力学,研究了 H3K27me3 在生理背景下的 TOR 调节。光合作用或外源性葡萄糖可以激活 TOR,其通过在光4、15、21 下 pS6K1(T449) 的磷酸化状态进行监测(图 4)。1d,e)。拟南芥幼苗在无糖液体培养基中发芽和生长,在生长发育中自然进入可逆静止状态,在发芽后 3 至 7 天,TOR 活性降低4,15(图 D)。在此期间,H3K27me3 水平在含糖培养基中保持不变(扩展数据图 1)。1e),但在糖饥饿阶段逐渐减少,与不活跃的 TOR 和停滞的幼苗发育有关。葡萄糖添加在 2 小时内迅速刺激了 TOR 活性和从头 H3K27me3 积累,表明染色质状态的重编程。一致地,在葡萄糖刺激期间,Torin2 (10 μM) 阻断了 H3K27me3 水平的恢复(图 D)。1e)。
To ensure TOR kinase specificity and to differentiate TOR regulation of H3K27me3 dynamics from effects of pS6K1(T449) (Fig. 1a), seedlings were treated with structurally distinct chemical inhibitors that target TOR or S6K1 (Fig. 1f). All five of the TOR inhibitors, but not the S6K1 inhibitors2,4, resulted in significant global H3K27me3 reduction. TOR kinase inhibition by Torin2 or AZD8055 led to similar genome-wide reduction of H3K27me3 occupancy as in tor-es mutants, as indicated by ChIP-Rx–seq analyses quantified by heat map, average density plot and genome browser view (Fig. 1g and Extended Data Fig. 2a–d and Supplementary Table 2). In seven-day-old glucose-starved seedlings, the conserved components of TOR complex 1 (TORC1), RAPTOR1 (RAP1) and LST8-1 (refs. 2,3,4,5), mediated S6K phosphorylation of pRPS6(S237) and pRPS6(S240), which were differentially blocked in the rap1 and lst8-1 mutants but not in rap2 or lst8-2 mutants24,25,26. However, rap1 and lst8-1 mutants could still substantially restore H3K27me3 levels after 6 h of stimulation with 25 mM glucose (Fig. 1d–g and Extended Data Fig. 2e). These results were consistent with different TOR activity thresholds modulating pS6K1(T449) and H3K27me3 revealed by the chemical inhibitor screens (Fig. 1a,f,g and Extended Data Fig. 2a), and suggest differential TOR kinase activation of distinct substrates beyond the canonical TORC1 (refs. 24,25).
为了确保 TOR 激酶的特异性并区分 H3K27me3 动力学的 TOR 调节与 pS6K1 (T449) 的影响(图 D)。1a),用靶向 TOR 或 S6K1 的结构不同的化学抑制剂处理幼苗(图 D)。所有 5 种 TOR 抑制剂,但不是 S6K1 抑制剂2,4,导致整体 H3K27me3 显着降低。Torin2 或 AZD8055 的 TOR 激酶抑制导致 H3K27me3 在全基因组的占有率降低,如 ChIP-Rx-seq 分析所示,通过热图、平均密度图和基因组浏览器视图进行量化(图 D)。1g 和扩展数据图2a-d 和补充表 2)。在 7 日龄的葡萄糖饥饿幼苗中,TOR 复合物 1 (TORC1)、RAPTOR1 (RAP1) 和 LST8-1 (参考文献.2,3,4,5) 介导 pRPS6 (S237) 和 pRPS6 (S240) 的 S6K 磷酸化,它们在 rap1 和 lst8-1 突变体中被差异阻断,但在 rap2 或 lst8-2 突变体中被差异阻断24,25,26。然而,rap1 和 lst8-1 突变体在用 25 mM 葡萄糖刺激 6 小时后仍能显着恢复 H3K27me3 水平(图 D)。1d-g 和扩展数据图这些结果与化学抑制剂筛选揭示的调节 pS6K1(T449) 和 H3K27me3 的不同 TOR 活性阈值一致(图 2)。1a,f,g 和扩展数据图2a),并表明经典 TORC1 之外不同底物的 TOR 激酶激活差异 (refs.24,25)。
TOR kinase promotes nuclear FIE dynamics
TOR 激酶促进核 FIE 动力学
The rapid increase in H3K27me3 stimulated by glucose–TOR signalling and the genome-wide reduction of H3K27me3 in tor-es seedlings suggested that TOR might regulate the activity of nuclear PRC2. In postembryonic development, Arabidopsis PRC2 comprises four evolutionarily conserved core subunits, including a methyltransferase (CURLY LEAF (CLF) or SWINGER (SWN)), a scaffold protein (EMBRYONIC FLOWER2 (EMF2) or VERNALIZATION2 (VRN2)), the histone-binding protein MULTICOPY-SUPPRESSOR-OF-IRA1 (MSI1) and FIE, which contains a WD40 structural fold6,7,8,10,11,12 (Extended Data Fig. 3a). We first examined the transcript and protein levels of PRC2 core components in tor-es or Torin2-treated seedlings; only CLF transcript and EMF2 protein were partially reduced (Extended Data Fig. 3b,c). We postulated that TOR might interact with and phosphorylate the core components and activate PRC2. We used pupylation-based interaction tagging27 (PUP-IT), a proximity-tagging system, to identify candidate substrates of TOR. We generated a haemagglutinin-tagged protein fusion of the bacterial Pup ligase (PafA) and the C-terminal kinase domain of TOR (TOR-C–PafA–HA) and co-expressed this with Flag-tagged Pup(E) and MYC-tagged PRC2 components in mesophyll protoplasts (Fig. 2a). FIE was specifically pupylated by TOR-C–PafA–HA (Fig. 2b). FIE is an essential and unique component of PRC2 complex, as the fie null mutant abolishes global H3K27me3 and limits organogenesis beyond germination12. The specific interaction between TOR and FIE in vivo was confirmed by co-immunoprecipitation, which was significantly enhanced after 2 h of stimulation with 5–50 mM glucose (Fig. 2c and Extended Data Fig. 4a). FIE was directly phosphorylated in vitro by the TOR kinase immunoprecipitated from Arabidopsis plants4; the phosphorylation reaction was specifically and completely inhibited by Torin2 (Fig. 2d).
葡萄糖-TOR 信号刺激的 H3K27me3 的快速增加和 tor-es 幼苗中 H3K27me3 的全基因组减少表明 TOR 可能调节核 PRC2 的活性。在胚胎后发育中,拟南芥 PRC2 包含四个进化上保守的核心亚基,包括甲基转移酶(卷曲叶 (CLF) 或 SWINGER (SWN))、支架蛋白(胚胎FLOWER2 (EMF2) 或VERNALIZATION2 (VRN2))、组蛋白结合蛋白 MULTICOPY-SUPPRESSOR-OF-IRA1 (MSI1) 和 FIE,其中包含 WD40 结构折叠 6,7,8,10,11,12(扩展数据图 D)。我们首先检测了 tor-es 或 Torin2 处理的幼苗中 PRC2 核心成分的转录和蛋白水平;只有 CLF 转录本和 EMF2 蛋白部分减少(扩展数据图 1)。3b,c)。我们假设 TOR 可能与核心组分相互作用并磷酸化并激活 PRC2。我们使用基于泡蛋白化的交互标记27 (PUP-IT),一种邻近标记系统,来识别 TOR 的候选底物。我们产生了细菌 Pup 连接酶 (PafA) 和 TOR (TOR-C-PafA-HA) 的 C 端激酶结构域的血凝素标记蛋白融合,并将其与 Flag 标记的 Pup(E) 和 MYC 标记的 PRC2 成分在叶肉原生质体中共表达(图 D)。FIE 被 TOR-C-PafA-HA 特异性化(图 2)。FIE 是 PRC2 复合物的重要组成部分,因为 fie 无效突变体消除了整体 H3K27me3 并限制了发芽后的器官发生12。 免疫共沉淀证实了体内 TOR 和 FIE 之间的特异性相互作用,在用 5-50 mM 葡萄糖刺激 2 小时后,这种相互作用显着增强(图 D)。2c 和扩展数据图FIE 在体外被拟南芥植物免疫沉淀的 TOR 激酶直接磷酸化4;磷酸化反应被 Torin2 特异性完全抑制(图 D)。2d)。
图 2:直接 TOR 磷酸化促进 FIE 从细胞质到细胞核的易位。
a, Schematic of the PUP-IT proximity-tagging system. PafA, a bacterial Pup ligase, is fused to the C terminus (residues 1226–2480) of TOR (TOR-C) (bait). TOR-C–PafA–HA joins 3×Flag–Pup(E) to lysine residues of interacting proteins (prey). The subunits of PRC2 were screened for interaction with TOR-C–PafA–HA by pupylation. b, Specific interaction between FIE and TOR. Immunoblot analysis of PUP-IT screening of MYC-tagged PRC2 components with TOR-C–PafA–HA. c, TOR interacts with FIE in vivo. Co-immunoprecipitation (Co-IP) of Flag-tagged FIE with TOR from seedlings crosslinked with formaldehyde. Non-transgenic WT seedlings served as the control. d, TOR directly phosphorylates FIE in vitro. Top, phosphorylation of His–FIE by immunoprecipitated endogenous TOR is shown by autoradiography. Bottom, Coomassie blue staining, showing protein loading. Torin2 specifically inhibits TOR kinase. e, FIE S14 is phosphorylated by TOR in vivo, as shown by tandem mass spectrometry. f, Critical TOR phosphorylation sites in FIE. In vitro TOR kinase assays were conducted with FIE and mutants. ΔN, N terminus deletion. SSTS/AAAA, the phosphorylation site mutant. g, Detection of FIE phosphorylation in vivo. CIP, calf Intestinal alkaline phosphatase. Values are the relative level of phosphorylation over the amount of immunoprecipitated Flag–GFP–FIE (labelled Flag–FIE), with mock treatment set as 1.0. h, Glucose enhances pFIE(S14) levels. Immunoblot analysis of pFIE(S14) in immunoprecipitated Flag–GFP–FIE from 7-day starved and 25 mM glucose-stimulated (2 h) WT and tor-es plants. i,j, Confocal images of GFP–FIE (i) and GFP–FIE(SSTS/AAAA) (j) in leaf primordia and roots. WT seedlings that were untreated (control) or treated with 10 μM Torin2 or AZD8055, or tor-es seedlings (treated with 10 μM oestradiol for 3 days). Inset, GFP detected in the root elongation zone. Scale bars, 25 μm. Images are representative of six seedlings from three biological repeats. Data in b–g,h are representative of three biological replicates each. The samples derive from the same experiment and gels and blots were processed in parallel.
a,PUP-IT 接近标记系统示意图。PafA 是一种细菌 Pup 连接酶,与 TOR (TOR-C)(诱饵)的 C 末端(残基 1226-2480)融合。TOR-C-PafA-HA 将 3×Flag-Pup(E) 连接到相互作用蛋白(猎物)的赖氨酸残基。通过化泡蛋白化筛选 PRC2 的亚基与 TOR-C-PafA-HA 的相互作用。b, FIE 和 TOR 之间的特定交互。使用 TOR-C-PafA-HA 对 MYC 标记的 PRC2 成分进行 PUP-IT 筛选的免疫印迹分析。c,TOR 在体内与 FIE 相互作用。来自甲醛交联幼苗的 Flag 标记的 FIE 与 TOR 的免疫共沉淀 (Co-IP)。非转基因 WT 幼苗作为对照。d,TOR 在体外直接磷酸化 FIE。上图,放射自显影显示免疫沉淀的内源性 TOR 对 His-FIE 的磷酸化。下图,考马斯蓝染色,显示蛋白质上样。Torin2 特异性抑制 TOR 激酶。e,FIE S14 在体内被 TOR 磷酸化,如串联质谱所示。f,FIE 中的关键 TOR 磷酸化位点。用 FIE 和突变体进行体外 TOR 激酶测定。ΔN,N 末端缺失。SSTS/AAAA,磷酸化位点突变体。g,体内 FIE 磷酸化检测。CIP, 小牛 肠道碱性磷酸酶。值是免疫沉淀的 Flag-GFP-FIE(标记为 Flag-FIE)量上的磷酸化相对水平,模拟处理设置为 1.0。h,葡萄糖增强 pFIE(S14) 水平。免疫印迹分析来自饥饿 7 天和 25 mM 葡萄糖刺激(2 小时)WT 和 tor-es 植物的免疫沉淀 Flag-GFP-FIE 中的 pFIE(S14)。 i,j,叶原基和根中 GFP-FIE (i) 和 GFP-FIE(SSTS/AAAA) (j) 的共聚焦图像。未处理(对照)或用 10 μM Torin2 或 AZD8055 处理的 WT 幼苗,或 tor-es 幼苗(用 10 μM 雌二醇处理 3 天)。插图,在根伸长区检测到 GFP。比例尺,25 μm。图像代表了来自三个生物重复序列的六棵幼苗。b-g,h 中的数据分别代表 3 个生物学重复。样品来源于同一实验,凝胶和印迹平行处理。
To identify the TOR phosphorylation site in FIE, we analysed phosphorylation sites from in vitro TOR kinase assays by liquid chromatography–tandem mass spectrometry (LC–MS/MS). We identified four main phosphorylation sites within the N-terminal domain of FIE: S10, S14, T16 and S18 (Extended Data Fig. 4b–f). Structural modelling of FIE based on embryonic ectoderm development protein (EED), the human orthologue of FIE, identified the flexible and serine- and threonine-rich N-terminal domain as a potential target for phosphorylation11,28 (Extended Data Fig. 4g). The phosphorylation of pS14, pT16 and pS18 in endogenous FIE was validated by LC–MS/MS analyses in glucose-stimulated seedlings (Fig. 2e and Extended Data Fig. 4b). Since S14A was the only single mutation in FIE that exhibited significantly reduced phosphorylation in the TOR kinase assay, the other phosphorylation sites probably have partially redundant and cooperative functions (Extended Data Fig. 4h). The N-terminal 34-residue deletion (ΔN) or the quadruple phosphorylation site mutant (SSTS to AAAA) of FIE was barely phosphorylated by TOR (Fig. 2f and Extended Data Fig. 4h). The key phosphorylation sites in FIE proteins are largely conserved among diverse flowering plants, including Arabidopsis, Brassica, soybean, rice, maize and wheat, and possibly in fly (ESC) and human (EED) orthologues of plant FIE29 (Extended Data Fig. 5a,b).
为了鉴定 FIE 中的 TOR 磷酸化位点,我们通过液相色谱-串联质谱 (LC-MS/MS) 分析了体外 TOR 激酶测定的磷酸化位点。我们在 FIE 的 N 端结构域内确定了四个主要磷酸化位点:S10、S14、T16 和 S18(扩展数据图 .4b-f)。基于胚胎外胚层发育蛋白 (EED)(FIE 的人类直系同源物)的 FIE 结构模型确定了灵活且富含丝氨酸和苏氨酸的 N 末端结构域作为磷酸化的潜在靶标11,28(扩展数据图 1)。4g). 通过葡萄糖刺激幼苗中的 LC-MS/MS 分析验证了内源性 FIE 中 pS14、pT16 和 pS18 的磷酸化(图 2)。2e 和扩展数据图由于 S14A 是 FIE 中唯一在 TOR 激酶测定中表现出磷酸化显著降低的单一突变,因此其他磷酸化位点可能具有部分冗余和协同功能(扩展数据图 .FIE 的 N 端 34 个残基缺失 (ΔN) 或四重磷酸化位点突变体(SSTS 到 AAAA)几乎没有被 TOR 磷酸化(图 D)。2f 和扩展数据图FIE 蛋白中的关键磷酸化位点在很大程度上保存在不同的开花植物中,包括拟南芥、芸苔属、大豆、水稻、玉米和小麦,并且可能在植物 FIE29 的果蝇 (ESC) 和人 (EED) 直系同源物中(扩展数据图 2)。5a,b)。
To functionally validate and quantify endogenous FIE phosphorylation, we generated a phosphopeptide-specific antibody targeting the phosphorylated form of FIE at S14, which was the most highly enriched and confident site validated by LC–MS/MS analyses in vitro and in planta (Fig. 2e and Extended Data Fig. 4b,d). This antibody specifically recognized the TOR-phosphorylated FIE, but not the S14A mutant (Extended Data Fig. 5c). Immunoprecipitation from seedlings did not detect in vivo phosphorylation of FIE at S14 (pFIE(S14)) following treatment with calf intestine phosphatase. Moreover, pFIE(S14) was largely abolished in seedlings treated with Torin2 or in tor-es plants (Fig. 2g). Immunoblot analysis showed that after starvation, pFIE(S14) was significantly induced within 2 h by 5–50 mM glucose; this effect was abolished in 7-day-old tor-es seedlings (Fig. 2h and Extended Data Fig. 5d). These results demonstrated that the N terminus of FIE is directly phosphorylated in glucose–TOR signalling.
为了对内源性 FIE 磷酸化进行功能验证和定量,我们制备了一种磷酸肽特异性抗体,靶向 S14 位点的磷酸化 FIE,这是在体外和植物中通过 LC-MS/MS 分析验证的最富集和最可靠的位点(图 D)。2e 和扩展数据图4b,d)。该抗体特异性识别 TOR 磷酸化的 FIE,但不识别 S14A 突变体(扩展数据图 .5c). 幼苗免疫沉淀未检测到小牛肠磷酸酶处理后 FIE 在 S14 (pFIE(S14)) 的体内磷酸化。此外,pFIE(S14) 在用 Torin2 处理的幼苗或 tor-es 植物中基本被消除(图 D)。免疫印迹分析显示,饥饿后 pFIE(S14) 在 2 小时内被 5–50 mM 葡萄糖显著诱导;这种影响在 7 日龄的 tor-es 幼苗中被消除(图 D)。2h 和扩展数据图这些结果表明,FIE 的 N 末端在葡萄糖-TOR 信号传导中被直接磷酸化。
We next explored how TOR phosphorylation regulates FIE function. Immunoblot analysis showed that the stability of GFP–FIE was not affected in tor-es or Torin2-treated plants (Extended Data Fig. 5e). Complexes formed by recombinant Arabidopsis PRC2 with the wild-type or the phosphorylation-site SSTS/AAAA mutant of FIE exhibited similar H3K27me3 methyltransferase activity in vitro (Extended Data Fig. 5f). Cytoplasmic FIE–GFP has been reported to interact exclusively with MEDEA (MEA) methyltransferase and not with CLF or SWN in inflorescences. It was postulated that the cytoplasmic FIE–GFP might have non-nuclear functions beyond chromatin methylation30. We tested the possibility that TOR phosphorylation promotes the cytoplasm-to-nucleus translocation of GFP–FIE to activate PRC2 (Extended Data Fig. 6a). Distinct from the predominant nuclear localization in control plants grown in sugar-containing nutrient medium, GFP–FIE was detected primarily in the cytoplasm in differentiating leaf primordia and root elongation and meristem zones in tor-es plants or following treatment of 5-day seedlings with Torin2 or AZD8055 (Fig. 2i and Extended Data Fig. 6b). However, the nuclear localization of other PRC2 components in roots—GFP–CLF, SWN–GFP, EMF2–GFP and MSI1–GFP—was not altered by Torin2 treatment (Extended Data Fig. 6c). Notably, the mutation of four TOR-phosphorylated sites (SSTS/AAAA) compromised the nuclear translocation of GFP–FIE in protoplasts and in transgenic plants without affecting the protein level (Fig. 2j and Extended Data Fig. 6b,d,e). The ratio of cytoplasmic to nuclear GFP–FIE (C:N ratio) increased quantitatively in the root elongation zone during the sugar starvation phase and decreased rapidly upon glucose treatment after five days of starvation (Extended Data Fig. 6f–j). Time-lapse live imaging showed that glucose rapidly stimulated real-time nuclear translocation of GFP–FIE within 2–4 h, coinciding with the increase in H3K27me3 level (Fig. 1e and Supplementary Video). In contrast to the conventional view of a preformed nuclear PRC2 with FIE as a key static component interacting with CLF or SWN and binding to H3K27me3 in the nucleus6,7,8,10,11,12,30, our findings revealed that FIE serves as a molecular bridge, directly linking glucose–TOR signalling with PRC2-regulated H3K27me3 dynamics and gene silencing.
接下来,我们探讨了 TOR 磷酸化如何调节 FIE 功能。免疫印迹分析表明,GFP-FIE 的稳定性在 tor-es 或 Torin2 处理的植物中不受影响(扩展数据图 .重组拟南芥 PRC2 与 FIE 的野生型或磷酸化位点 SSTS/AAAA 突变体形成的复合物在体外表现出相似的 H3K27me3 甲基转移酶活性(扩展数据图 D)。据报道,细胞质 FIE-GFP 仅与 MEDEA (MEA) 甲基转移酶相互作用,而不与花序中的 CLF 或 SWN 相互作用。据推测,细胞质 FIE-GFP 可能具有除染色质甲基化之外的非核功能30。我们测试了 TOR 磷酸化促进 GFP-FIE 细胞质到细胞核易位以激活 PRC2 的可能性(扩展数据图 1)。与在含糖营养培养基中生长的对照植物中的主要核定位不同,GFP-FIE 主要在 tor-es 植物的叶原基和根伸长和分生组织区的细胞质中检测到,或在用 Torin2 或 AZD8055 处理 5 天幼苗后(图 D)。图2i 和扩展数据图然而,其他 PRC2 组分在根中的核定位——GFP-CLF、SWN-GFP、EMF2-GFP 和 MSI1-GFP——并没有被 Torin2 处理改变(扩展数据图 D)。值得注意的是,四个 TOR 磷酸化位点 (SSTS/AAAA) 的突变损害了原生质体和转基因植物中 GFP-FIE 的核转位,而不会影响蛋白质水平(图 D)。2j 和扩展数据图6b,d,e)。 在糖饥饿阶段,细胞质与核 GFP-FIE 的比率(C:N 比率)在根伸长区数量上增加,并在饥饿 5 天后葡萄糖处理后迅速下降(扩展数据图 1)。6f-j)。延时实时成像显示,葡萄糖在 2-4 小时内迅速刺激了 GFP-FIE 的实时核转位,与 H3K27me3 水平的增加相吻合(图 D)。1e 和补充视频)。与传统观点相比,以 FIE 为关键静态成分的预制核 PRC2 与 CLF 或 SWN 相互作用并与细胞核中的 H3K27me3 结合 6,7,8,10,11,12,30,我们的研究结果显示,FIE 充当分子桥,直接将葡萄糖-TOR 信号与 PRC2 调节的 H3K27me3 动力学和基因沉默联系起来。
TOR phosphorylation of FIE controls development
FIE 的 TOR 磷酸化控制发育
To elucidate the molecular functions of the phosphorylated FIE by TOR kinase, we introduced GFP–FIE with or without the phosphorylation site mutation SSTS/AAAA, under the control of the FIE promoter, into heterozygous fie/+ plants. Homozygous GFP-FIE/fie and SSTS/AAAA/fie plants were selected for genome-wide H3K27me3 and transcriptome analyses. Conditional fie mutants that bypass embryonic lethality were also generated to provide a parallel comparison with SSTS/AAAA/fie (Extended Data Fig. 7a–d). The oestradiol-inducible fie-amiR-es transgenic lines eliminated FIE protein and exhibited consistent aberrant small, narrow and curled true leaves sharing some features with the occasional FIE co-suppressed plants31 (Extended Data Fig. 7b–d). The fie null mutant seeds generated with the heterozygous fie/+ cdka;1/+ pollen forms only undifferentiated callus-like structures after germination and blocks further postembryonic development12. The SSTS/AAAA/fie and fie-amiR-es mutants had a weaker effect and remained capable of postembryonic development and organogenesis powered by photosynthesis or exogenous glucose4, thus providing a valuable platform to investigate the physiological functions of the glucose–TOR–FIE–PRC2 signalling network.
为了阐明 TOR 激酶磷酸化 FIE 的分子功能,我们在 FIE 启动子的控制下,将有或没有磷酸化位点突变 SSTS/AAAA 的 GFP-FIE 引入杂合 FIE/+ 植物中。选择纯合 GFP-FIE/fie 和 SSTS/AAAA/fie 植物进行全基因组 H3K27me3 和转录组分析。还生成了绕过胚胎致死性的条件 fie 突变体,以提供与 SSTS/AAAA/fie 的平行比较(扩展数据图 1)。7a-d)。雌二醇诱导型 fie-amiR-es 转基因系消除了 FIE 蛋白,并表现出一致异常的小、窄和卷曲的真叶,与偶尔的 FIE 共抑制植物共享一些特征31(扩展数据图 3)。7b-d)。用杂合子 fie/+ cdka 生成的 fie 无效突变种子;1/+ 花粉在发芽后仅形成未分化的愈伤组织样结构,并阻止胚胎后进一步发育12。SSTS/AAAA/fie 和 fie-amiR-es 突变体的作用较弱,并且仍然能够在光合作用或外源葡萄糖4 的驱动下进行胚胎后发育和器官发生,从而为研究葡萄糖-TOR-FIE-PRC2 信号网络的生理功能提供了一个有价值的平台。
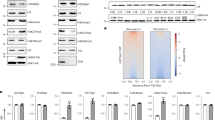
Quantitative immunoblot and ChIP-Rx–seq analyses revealed markedly decreased global H3K27me3 levels across the genome in the 14-day vegetative shoots of SSTS/AAAA/fie and fie-amiR-es mutants compared with those in WT and GFP-FIE/fie plants (Fig. 3a,b, Extended Data Fig. 7e,f and Supplementary Table 3). The SSTS/AAAA/fie mutant showed a larger reduction in H3K27me3 than those resulting from TOR inhibition, suggesting that the phosphorylation mutant of FIE was decoupled from other TOR-regulated processes, including DNA replication that could limit H3K27me3 dilution when PRC2 was inhibited2,3,4,5,15 (Fig. 1a and Extended Data Figs. 1b–d and 2b–d). As a parallel control, GFP–FIE expression in the fie null mutant restored the H3K27me3 landscape (Fig. 3a,b, Extended Data Fig. 7e,f and Supplementary Table 3). Genome-wide H3K27me3 target genes previously identified by ChIP analyses in 14-day to 20-day WT seedlings significantly overlapped with our data from quantitative ChIP-Rx–seq analyses in 14-day WT shoots12,32 (Extended Data Fig. 7g). Consistent with the genome-wide profiling, H3K27me3 occupancy was decreased at selected target loci with key developmental roles6,7,8,12,31,32 in the SSTS/AAAA/fie and fie-amiR-es mutants (Fig. 3c).
定量免疫印迹和 ChIP-Rx-seq 分析显示,与 WT 和 GFP-FIE/fie 植物相比,SSTS/AAAA/fie 和 fie-amiR-es 突变体的 14 天营养芽中整个基因组的整体 H3K27me3 水平显著降低(图 D)。3a,b,扩展数据图7e,f 和补充表 3)。SSTS/AAAA/fie 突变体显示 H3K27me3 的减少幅度大于 TOR 抑制导致的 H3K27me3 减少,这表明 FIE 的磷酸化突变体与其他 TOR 调节过程解耦,包括当 PRC2 被抑制时可以限制 H3K27me3 稀释的 DNA 复制 2,3,4,5,15(图 D)。1a 和扩展数据图1b-d 和 2b-d)。作为平行对照,fie 无效突变体中的 GFP-FIE 表达恢复了 H3K27me3 景观(图 D)。3a,b,扩展数据图7e,f 和补充表 3)。先前在 14 天至 20 天 WT 幼苗中通过 ChIP 分析鉴定的全基因组 H3K27me3 靶基因与我们在 14 天 WT 芽中来自定量 ChIP-Rx-seq 分析的数据显着重叠12,32(扩展数据图 1)。与全基因组分析一致,H3K27me3 在选定的靶基因座上降低,在 SSTS/AAAA/fie 和 fie-amiR-es 突变体中具有关键发育作用6,7,8,12,31,32 (图 D)。3c)。
图 3:靶向 FIE 磷酸化突变改变了整体 H3K27me3 景观,以重新编程转录组并破坏器官发生。
a, Reduction of global H3K27me3 levels in 14-day-old fie mutant plants. Metaplots show ChIP-Rx–seq read density of H3K27me3 ±2 kb from their peak summits. Chromatin immunoprecipitation with sequencing (ChIP–seq) data are normalized with an exogenous reference genome. b, Depletion of H3K27me3 in fie mutants. Heat map of H3K27me3 enrichment. The colour scale indicates RRPM surrounding the peak summit, obtained from the ChIP-Rx–seq data. c, The IGV browser view of H3K27me3 occupancy at PRC2 target genes. ACT2, a non-PRC2-target gene, serves as a negative control. d, Transcriptome reprogramming in 14-day fie mutants. The heat map shows transcriptomic changes of fie-amiR-es (versus WT) and SSTS/AAAA (versus GFP-FIE) from RNA-seq analyses. FC, fold change. e, Comparative expression analyses of key transcription factor genes in diverse developmental programmes. Heat map of RNA-seq data from triplicate biological samples prepared from WT, fie-amiR-es, GFP-FIE/fie and SSTS/AAAA/fie plants. The RNA expression data were normalized to the value in WT plants. f, The fie mutants display broad developmental aberrations. The fie-amiR-es and SSTS/AAAA/fie mutants exhibit small, narrow and curled leaves and flower prematurely at 21 days. The SSTS/AAAA/fie mutants develop thin, short and terminal inflorescence with aborted flower buds, narrow, split and twisted sepals and petals, and enlarged carpel, as well as short, bulged and contorted siliques (at 30 days). g, GUS reporter detection in 14-day-old GFP-FIE/fie and SSTS/AAAA/fie mutant plants transfected with pKNAT1-GUS. Arrowheads indicate SAM. Three biological replicates, n > 10 plants per experiment. White scale bar, 10 mm; red scale bar, 1 mm.
a,降低 14 日龄 fie 突变植物中的整体 H3K27me3 水平。Meta 图显示 H3K27me3 的 ChIP-Rx-seq 读取密度从其峰值开始±2 kb。染色质免疫沉淀测序 (ChIP–seq) 数据使用外源参考基因组进行归一化。b, fie 突变体中 H3K27me3 的消耗。H3K27me3 富集的热图。色标表示峰顶周围的 RRPM,从 ChIP-Rx-seq 数据获得。c, H3K27me3 在 PRC2 靶基因上占有率的 IGV 浏览器视图。ACT2 是一种非 PRC2 靶基因,用作阴性对照。d, 14 天 fie 突变体中的转录组重编程。热图显示了 RNA-seq 分析中 fie-amiR-es (与 WT) 和 SSTS/AAAA (与 GFP-FIE) 的转录组变化。FC,换折。e,不同发育计划中关键转录因子基因的比较表达分析。来自 WT、fie-amiR-es、GFP-FIE/fie 和 SSTS/AAAA/fie 植物制备的一式三份生物样品的 RNA-seq 数据的热图。将 RNA 表达数据标准化为 WT 植物中的值。f, fie 突变体表现出广泛的发育畸变。fie-amiR-es 和 SSTS/AAAA/fie 突变体表现出小、窄和卷曲的叶子,并在 21 天时过早开花。SSTS/AAAA/fie 突变体长出薄、短和末端的花序,花芽流产,萼片和花瓣狭窄、分裂和扭曲,心皮增大,以及短、凸和扭曲的角果(在 30 天时)。 g, 转染 pKNAT1-GUS 的 14 日龄 GFP-FIE/fie 和 SSTS/AAAA/fie 突变株中的 GUS 报告基因检测。箭头表示 SAM。3 个生物学重复,每个实验 n > 10 株植物。白色鳞片,10 毫米;红色刻度条,1 毫米。
Transcriptome profiling by RNA-seq identified a total of 3,969 genes (|log2 fold change| ≥ 1; q ≤ 0.05, n = 3) that were dysregulated in the 14-day shoots of SSTS/AAAA/fie and fie-amiR-es plants (Fig. 3d and Supplementary Table 4). To uncover the biological functions of the TOR–FIE–PRC2 signalling network, we defined the putative direct target genes that were marked by H3K27me3 and upregulated in the SSTS/AAAA/fie mutant (Extended Data Fig. 8a and Supplementary Table 5). Gene ontology analysis of 1,081 (log2 fold change ≥ 1; q ≤ 0.05) TOR–FIE–PRC2 target genes revealed a notable enrichment for transcription factors and regulators controlling a broad spectrum of developmental programmes (Extended Data Fig. 8b and Supplementary Table 6). We identified 192 transcription factor genes (Supplementary Table 7), including master regulators with essential roles in stem cell identity, cell fate determination, patterning and developmental transitions, that were significantly upregulated and associated with reduction of H3K27me3 in SSTS/AAAA/fie and fie-amiR-es mutants (Fig. 3e and Supplementary Tables 5 and 7). In particular, the expression of shoot apical meristem (SAM)-specific transcription factors defining the indeterminant cell fate33 increased in the mutant plants. The abnormal expression of root quiescent centre and stem cell regulators34,35 were detected in the 14-day shoots. The key floral integrators controlling vegetative-to-reproductive transition were precociously derepressed at the vegetative seedling stage. We also observed the ectopic activation of floral meristem and floral homeotic genes36 in seedlings. Important transcription factor genes governing leaf development37,38 as well as shoot branching16,39 were also overexpressed in the SSTS/AAAA/fie and fie-amiR-es mutants (Fig. 3e, Extended Data Fig. 8c and Supplementary Tables 5 and 7).
通过 RNA-seq 进行的转录组分析共鉴定出 3,969 个基因 (|log2 倍变化|≥ 1;q ≤ 0.05,n = 3),它们在 SSTS/AAAA/fie 和 fie-amiR-es 植物的 14 天芽中失调(图 D)。3d 和补充表 4)。为了揭示 TOR-FIE-PRC2 信号网络的生物学功能,我们定义了由 H3K27me3 标记并在 SSTS/AAAA/fie 突变体中上调的推定直接靶基因(扩展数据图 .8a 和补充表 5)。1,081 的基因本体分析 (对数2 倍变化≥ 1;q ≤ 0.05) TOR-FIE-PRC2 靶基因揭示了控制广泛发育程序的转录因子和调节因子的显着富集(扩展数据图 .8b 和补充表 6)。我们鉴定了 192 个转录因子基因(补充表 7),包括在干细胞身份、细胞命运决定、模式化和发育转变中起重要作用的主调节因子,这些基因在 SSTS/AAAA/fie 和 fie-amiR-es 突变体中显著上调并与 H3K27me3 的减少相关(图 7)。3e 和补充表 5 和 7)。特别是,定义决定不确定细胞命运33 的茎尖分生组织 (SAM) 特异性转录因子的表达在突变植物中增加。在 14 d 的芽中检测到根静止中心和干细胞调节因子34,35 的异常表达。控制营养到生殖转变的关键花整合者在营养幼苗阶段早熟去抑制。 我们还观察到幼苗中花分生组织和花同源基因36 的异位激活。控制叶片发育的重要转录因子基因37,38 以及芽分枝16,39 也在 SSTS/AAAA/fie 和 fie-amiR-es 突变体中过表达(图 .3e,扩展数据图8c 和补充表 5 和 7)。
In 7-day seedlings grown in liquid medium and optimal for starvation and glucose, and in tor-es experiments4,15,21, quantitative PCR with reverse transcription (RT–qPCR) analysis showed that a subset of TOR–FIE–PRC2–H3K27me3 target genes were upregulated, and this was correlated with a reduction of H3K27me3, as shown by chromatin immunoprecipitation with quantitative PCR (ChIP–qPCR) analysis (Extended Data Fig. 8d,e). Consistently, in the sugar-starved 7-day seedlings, glucose initiated the repression of these genes, which was correlated with elevated H3K27me3 at 6 h (Extended Data Fig. 8f,g). Although target gene regulation by tor-es or glucose was more subtle in 7-day seedlings than in 14-day SSTS/AAAA/fie and fie-amiR-es mutant plants (Fig. 3e and Extended Data Fig. 8c), our findings suggest that the glucose–TOR–FIE–PRC2 signalling pathway controls target genes encoding key transcription factors via an epigenomic mechanism (Supplementary Table 7). Further research will be required to elucidate the detailed integration of distinct TOR signalling mechanisms in temporospatial modulation of epigenomic and transcriptomic controls of key regulators in diverse processes during different stages of postembryonic development at organ, tissue and single-cell levels.
在液体培养基中生长的 7 天幼苗中,最适合饥饿和葡萄糖,以及在 tor-es 实验 4,15,21 中,定量 PCR 与逆转录 (RT-qPCR) 分析显示 TOR-FIE-PRC2-H3K27me3 靶基因的一个子集上调,这与 H3K27me3 的减少相关,如染色质免疫沉淀与定量 PCR (ChIP-qPCR) 分析所示(扩展数据图 .8d,e)。一致地,在缺糖的 7 天幼苗中,葡萄糖启动了这些基因的抑制,这与 6 小时时 H3K27me3 升高相关(扩展数据图 D)。8f,g)。尽管 7 天幼苗中 tor-es 或葡萄糖的靶基因调控比 14 天 SSTS/AAAA/fie 和 fie-amiR-es 突变植物更微妙(图 .3e 和扩展数据图8c),我们的研究结果表明,葡萄糖-TOR-FIE-PRC2 信号通路通过表观基因组机制控制编码关键转录因子的靶基因(补充表 7)。需要进一步的研究来阐明不同的 TOR 信号机制在器官、组织和单细胞水平胚胎后发育不同过程中关键调节因子的表观基因组和转录组控制的时间空间调节中的详细整合。
The consequences of H3K27me3 and transcriptome reprogramming in SSTS/AAAA/fie plants were clearly manifested in the profound disruption of temporal and spatial orders of the innate genetic programmes guiding plant differentiation and development31,33,34,35,36,37,38,39,40. Of note, in 21- to 24-day-old plants covering broader developmental stages, the SSTS/AAAA/fie mutants displayed grossly aberrant developmental phenotypes, similar to those in fie-amiR-es lines, including early flowering, abnormalities in the size, morphology and patterning of leaves, flowers and siliques, reduced branching with miniature plant architecture and terminal flowers and infertility (Fig. 3f and Extended Data Fig. 7d), as well as enlarged SAM indicated by the pKNAT1:GUS reporter (Fig. 3g). The expression of GFP–FIE in the null fie mutant fully restored the size and morphology of organs, whole plant architecture, developmental timing and fertility (Fig. 3f and Extended Data Fig. 7d). These data indicate that TOR phosphorylation of FIE has a vital role in gating PRC2-mediated epigenomic reprogramming in diverse postembryonic developmental programmes throughout the plant life.
SSTS/AAAA/fie 植物中 H3K27me3 和转录组重编程的后果清楚地表现在指导植物分化和发育的先天遗传程序的时空秩序的深刻破坏31、33、34、35、36、37、38、39、40。值得注意的是,在涵盖更广泛发育阶段的 21 至 24 日龄植物中,SSTS/AAAA/fie 突变体表现出严重异常的发育表型,类似于 fie-amiR-es 品系中的表型,包括早期开花,叶、花和角果的大小、形态和图案异常,微型植物结构和顶花分枝减少以及不育(图 D)。3f 和扩展数据图7d) 以及 pKNAT1:GUS 报告基因指示的增大 SAM (图 D)。GFP-FIE 在无效突变体中的表达完全恢复了器官的大小和形态、全株结构、发育时间和生育能力 (图 .3f 和扩展数据图这些数据表明,FIE 的 TOR 磷酸化在整个植物生命周期的不同胚后发育计划中对 PRC2 介导的表观基因组重编程的门控中起着至关重要的作用。
Glucose–TOR–FIE signalling gates vernalization
葡萄糖-TOR-FIE 信号门春化
To further explore how this direct molecular link between glucose–TOR signalling and H3K27me3 dynamics is involved in a defined developmental programme, we studied vernalization-induced flowering. This process involves H3K27me3-mediated silencing of the floral transcription repressor gene FLOWERING LOCUS C (FLC) by prolonged cold treatment, which provides a paradigm to study environmentally regulated epigenetic dynamics during developmental phase transitions40,41,42,43. FLC was activated in the SSTS/AAAA/fie and fie-amiR-es mutants (Fig. 3e), and sugar accumulation was found to be associated with prolonged cold treatment and floral induction13,44,45, but its physiological importance remains unknown. To decipher the role of glucose–TOR signalling in vernalization-mediated floral transition, we showed that prolonged cold exposure significantly induced sustained glucose accumulation in the Col-FRIGIDA (FRI) line (Fig. 4a). The functional FRI allele confers the vernalization-dependent floral transition in Arabidopsis42. The elevation of endogenous TOR activity quantified by pS6K1(T449) closely matched the profile of intrinsic glucose surge during vernalization in FRI and was abolished by potent and specific TOR inhibitors, Torin2 or INK128 (1 μM) (Figs. 1f,g and 4b). These results suggest that glucose-induced TOR activation may regulate vernalization.
为了进一步探索葡萄糖-TOR 信号传导和 H3K27me3 动力学之间的这种直接分子联系如何参与明确的发育计划,我们研究了春化诱导的开花。该过程涉及 H3K27me3 介导的花转录抑制基因 FLOWERING LOCUS C (FLC) 通过长时间冷处理沉默,这为研究发育阶段转变过程中环境调节的表观遗传动力学提供了一种范例40,41,42,43。FLC 在 SSTS/AAAA/fie 和 fie-amiR-es 突变体中被激活(图 D)。3e),发现糖积累与长时间的冷处理和花诱导有关13,44,45,但其生理重要性仍然未知。为了破译葡萄糖-TOR 信号转导在春化介导的花转变中的作用,我们发现长时间的寒冷暴露显着诱导了 Col-FRIGIDA (FRI) 系的持续葡萄糖积累(图 1)。功能性 FRI 等位基因赋予拟南芥42 中春化依赖性的花转变。由 pS6K1 (T449) 量化的内源性 TOR 活性的升高与 FRI 中春化过程中内源性葡萄糖激增的特征密切相关,并被有效和特异性的 TOR 抑制剂 Torin2 或 INK128 (1 μM) 消除(图1f、g 和 4b)。这些结果表明,葡萄糖诱导的 TOR 激活可能调节春化。
图 4:葡萄糖-TOR-FIE 信号刺激春化介导的花转变。
a, Vernalization induces glucose accumulation in Col-FRI plants. NV, non-vernalized; V, vernalization days at 4 °C; T, post-cold days at 22 °C. n = 3 biological repeats. b, Vernalization stimulates TOR activity. TOR activity was indicated by the band shift of S6K1–HA, indicating S6K1 phosphorylation (pS6K1). c, TOR promotes FLC repression by vernalization. d, TOR inhibitors prevent vernalization-mediated repression of the FLC reporter. Histochemical staining of the pFLC:FLC-GUS (Ler-FRI) transgenic plant before and after vernalization without or with TOR inhibitors (1 μM Torin2 or INK128). Scale bar, 10 mm. e, TOR deficiency reduces H3K27me3 levels at FLC. Black boxes indicate exons and red boxes show regions selected for H3K27me3 ChIP–qPCR analyses. bp, base pairs. FLC-1 and FLC-2 are located in the nucleation and spreading regions of H3K27me3 at FLC, respectively. f, TOR is required for vernalization. Flowering time was analysed by counting rosette leaf numbers when bolting after vernalization. Data are mean ± s.d. n is the number of individual plants. g, Representative flowering phenotype of the indicated plant at bolting after vernalization. Scale bar, 10 mm. h, FIE phosphorylation increases during vernalization. Immunoblot analysis of total and phosphorylated FIE proteins after vernalization treatment. Values show the relative level of phosphorylated FIE over total immunoprecipitated Flag–FIE. The non-vernalized level is set as 1.0. i, Repression of FLC by vernalization is diminished in SSTS/AAAA/fie/Col-FRI plants. j, Vernalization-induced H3K27me3 levels are compromised at FLC during two silencing phases. c,i, Relative expression of FLC versus UBC21 was normalized to the non-vernalized level in control or fie/Col-FRI plants. e,j, Relative H3K27me3 versus H3 level was normalized to that of control or fie/Col-FRI plants under the non-vernalized condition. Data in c,e,i,j are mean ± s.d. from 3 biological replicates; two-way ANOVA with Tukey’s multiple comparisons test. f, One-way ANOVA with Tukey’s multiple comparisons test. Data in b,h are representative of three biological replicates each. The samples derive from the same experiment and gels and blots were processed in parallel.
To strengthen the molecular link between the glucose–TOR–FIE–PRC2 signalling relay and vernalization, we quantified FLC expression, PRC2-mediated H3K27me3 marks on the FLC chromatin and flowering time. FRI was introduced into tor-es, GFP-FIE/fie and SSTS/AAAA/fie mutants for comparative analyses. Cold-induced repression of FLC was impaired by specific TOR inhibitors and in the tor-es mutant (Fig. 4c), whereas the activation of VERNALIZATION INSENSITIVE3 (VIN3) was unaffected during the same cold exposure (Extended Data Fig. 9a). To monitor the FLC expression dynamics and locations in intact plants, the transgenic pFLC:FLC-GUS (Ler-FRI) line carrying the FLC promoter and a GUS translational fusion was visualized by histochemical staining. Consistent with the RNA expression results, the reduction of FLC reporter expression in the SAM and vasculature upon vernalization was prevented by TOR inhibitors (Fig. 4d).
为了加强葡萄糖-TOR-FIE-PRC2 信号传递和春化之间的分子联系,我们量化了 FLC 表达、PRC2 介导的 FLC 染色质上的 H3K27me3 标记和开花时间。将 FRI 引入 tor-es 、 GFP-FIE/fie 和 SSTS/AAAA/fie 突变体中进行比较分析。低温诱导的 FLC 抑制受到特异性 TOR 抑制剂和 tor-es 突变体的损害(图 .4c),而 VERNALIZATION INSENSITIVE3 (VIN3) 的激活在相同的寒冷暴露期间不受影响(扩展数据图 1)。为了监测完整植物中 FLC 的表达动力学和位置,通过组织化学染色观察携带 FLC 启动子和 GUS 翻译融合的转基因 pFLC:FLC-GUS (Ler-FRI) 系。与 RNA 表达结果一致,TOR 抑制剂阻止了春化后 SAM 和脉管系统中 FLC 报告基因表达的降低(图 D)。4d)。
Cold-induced silencing of FLC by PRC2 was characterized by two phases: the initial nucleation and the propagation of H3K27me3 on the FLC locus42. We found that both phases were impaired in tor-es or by TOR inhibitors (Fig. 4e and Extended Data Fig. 9b). In addition, plants with conditional TOR deficiency during vernalization exhibited delayed flowering time after returning to ambient temperature with full TOR activity (Fig. 4f,g). These results demonstrate that FLC repression during vernalization requires TOR activity. Prolonged cold treatment markedly increased the accumulation of glucose (Fig. 4a), pS6K1 (Fig. 4b), pFIE(S14), total FIE protein level (Fig. 4h) and the quantitative nuclear localization of GFP–FIE (Extended Data Fig. 9c,d), which prompted the initial nucleation of H3K27me3 (refs. 4,40,42,43). The phosphorylation and nuclear localization of FIE were sustained even after the large glucose surge declined after 30 days of vernalization, presumably owing to continuous energy expenditure with limited photosynthesis under cold treatment4,46. The steady-state glucose level and TOR activity, and FIE phosphorylation and localization resumed to support normal and dynamic photosynthesis-driven glucose–TOR–FIE signalling for the maintenance and spreading of H3K27me3 after plants were returned to warm temperature4,40,42,43,46. The differential dynamics of pS6K1 and pFIE dephosphorylation during vernalization (Fig. 4b,h) suggested that distinct glucose–TOR substrates for divergent downstream signalling processes may exhibit different kinetics and regulation. Vernalization-mediated FLC repression was diminished during the cold exposure but recovered after cold in the SSTS/AAAA/fie mutant (Fig. 4i), resembling the vernalization-defective vin3, vrn2 and vrn5 mutants40,42. Consistently, the nucleation and spreading of H3K27me3 enrichment at FLC in response to vernalization were abolished in the SSTS/AAAA/fie mutant plants (Fig. 4j and Extended Data Fig. 9e). Together, these data indicate that specific FIE phosphorylation by glucose–TOR signalling modulate vernalization-mediated floral transition.
PRC2 对 FLC 的冷诱导沉默具有两个阶段:H3K27me3 在 FLC 基因座42 上的初始成核和繁殖。我们发现这两个阶段在 tor-es 或 TOR 抑制剂中都受损(图4e 和扩展数据图此外,春化过程中条件性 TOR 缺陷的植物在恢复到具有完全 TOR 活性的环境温度后表现出延迟的开花时间(图 D)。4f,g)。这些结果表明,春化过程中的 FLC 抑制需要 TOR 活性。长时间的冷处理显着增加了葡萄糖的积累(图 D)。4a)、pS6K1(图4b)、pFIE(S14)、总 FIE 蛋白水平 (图 .4h) 和 GFP-FIE 的定量核定位(扩展数据图 .9c,d),这促使 H3K27me3 的初始成核(参考文献。4,40,42,43)。即使在春化 30 天后大葡萄糖激增下降后,FIE 的磷酸化和核定位仍能持续,这可能是由于在冷处理下光合作用有限的情况下持续的能量消耗 4,46。稳态葡萄糖水平和 TOR 活性以及 FIE 磷酸化和定位恢复,以支持正常和动态光合作用驱动的葡萄糖-TOR-FIE 信号传导,在植物恢复到温暖温度后维持和扩散 H3K27me3 4,40,42,43,46。春化过程中 pS6K1 和 pFIE 去磷酸化的差异动力学(图 D)。 4b,h)表明用于不同下游信号传导过程的不同葡萄糖-TOR 底物可能表现出不同的动力学和调节。春化介导的 FLC 抑制在寒冷暴露期间减弱,但在 SSTS/AAAA/fie 突变体中寒冷后恢复(图 D)。4i),类似于春化缺陷的 vin3、vrN2 和 vrn5 突变体40,42。一致地,在 SSTS/AAAA/fie 突变植物中,响应春化而在 FLC 处富集的 H3K27me3 成核和扩散被消除(图 D)。4j 和扩展数据图总之,这些数据表明葡萄糖-TOR 信号传导的特异性 FIE 磷酸化调节春化介导的花转变。
Discussion 讨论
Here we report a direct molecular link between glucose–TOR signalling and PRC2 regulation. TOR kinase, activated by glucose derived from local or systemic carbon sources, phosphorylates FIE at S14 and other cooperative sites (S10, T16 and S18) and promotes its translocation from the cytoplasm to the nucleus to enhance PRC2 activity. In the nucleus, the active FIE–PRC2 complex is probably recruited by transcription repressors, cis-regulatory elements and non-coding RNAs to deposit H3K27me3 for silencing of key transcription factor genes that modulate cell fate determination, developmental transitions and organ patterning in Arabidopsis6,7,8,10,40,42,43,47,48,49. We propose that this signalling axis serves as a nutritional checkpoint for regulating PRC2 activity throughout key differentiation phases and processes during plant postembryonic development (Figs. 3 and 4 and Extended Data Fig. 10). Notably, there is a recent report of TOR regulation of H3K27me3 target genes that are activated by stress conditions beyond normal developmental programmes or vernalization50.
在这里,我们报道了葡萄糖-TOR 信号传导与 PRC2 调节之间的直接分子联系。TOR 激酶被来自局部或全身碳源的葡萄糖激活,在 S14 和其他合作位点 (S10、T16 和 S18) 磷酸化 FIE,并促进其从细胞质转位到细胞核以增强 PRC2 活性。在细胞核中,活性 FIE-PRC2 复合物可能被转录抑制因子、顺式调节元件和非编码 RNA 募集,以沉积 H3K27me3 以沉默调节拟南芥 6,7,8,10,40,42,43,47,48,49 的关键转录因子基因.我们提出,这个信号轴是一个营养检查点,在植物胚后发育过程中的关键分化阶段和过程中调节 PRC2 活性(图图 3 和 4 以及扩展数据图值得注意的是,最近有一份关于 H3K27me3 靶基因的 TOR 调节的报道,这些靶基因被超出正常发育程序或春化的应激条件激活50。
A notable feature of the postembryonic growth and development in plants is the indeterminate stem cell niche in the meristems, which continuously supply new cells for root, leaf, stem, flower and fruit organogenesis beyond the simple embryo designed for nutrient storage and dormancy12,33,34. Our genome-wide analyses define diverse master transcription factors as target genes of the glucose–TOR–PRC2 signalling network and key to major developmental programmes (Figs. 3 and 4, Extended Data Figs. 8 and 10 and Supplementary Table 7). This glucose-stimulated epigenomic network promotes faithful differentiation and developmental transitions, including stem cell to meristem or primordium switch, juvenile to adult organogenesis, axillary meristem to branching, and reproductive processes from flowering to silique development (Extended Data Fig. 10a). This molecular switch ensures that the proliferation and differentiation processes during organ growth and patterning in time and space are integrated and coordinated with adequate nutrient and energy support. Crucially, under the prolonged cold condition, the glucose-activated TOR–FIE–PRC2 relay overrides the default vegetative programme by silencing the floral repressor FLC, which may suggest how sugars support the vernalization-induced flowering44 (Extended Data Fig. 10b). These molecular connections provide a long-sought mechanistic explanation for how glucose signalling regulates multiple developmental processes in plants. This study advances a conceptual understanding of how multicellular organisms transmit systemic nutrient information to remodel global chromatin states and modulate local cellular chromatin regulators, thus orchestrating the transcriptional landscape that is central to cell fate regulation in diverse developmental programmes.
植物胚后生长发育的一个显着特征是分生组织中不确定的干细胞生态位,除了为营养储存和休眠而设计的简单胚胎之外,它不断为根、叶、茎、花和果实的器官发生提供新细胞12,33,34。我们的全基因组分析将不同的主转录因子定义为葡萄糖-TOR-PRC2 信号网络的靶基因,并且是主要发育计划的关键(图3 和 4,扩展数据图8 和 10 以及补充表 7)。这种葡萄糖刺激的表观基因组网络促进忠实的分化和发育转变,包括干细胞到分生组织或原基的转换、幼年到成体器官发生、腋生组织到分枝,以及从开花到角细胞发育的生殖过程(扩展数据图 .这种分子开关确保器官生长和时间和空间模式化过程中的增殖和分化过程与足够的营养和能量支持相结合和协调。至关重要的是,在长时间的寒冷条件下,葡萄糖激活的 TOR-FIE-PRC2 中继通过沉默花抑制因子 FLC 来覆盖默认的营养程序,这可能表明糖如何支持春化诱导的开花44 (扩展数据图 .这些分子连接为葡萄糖信号传导如何调节植物的多个发育过程提供了一个长期寻求的机制解释。 这项研究推进了对多细胞生物如何传递全身营养信息以重塑整体染色质状态和调节局部细胞染色质调节因子的概念理解,从而协调对不同发育计划中细胞命运调控至关重要的转录景观。
Methods 方法
Plasmid constructs and the generation of transgenic plants
质粒构建体和转基因植物的产生
The pFIE:Flag-GFP-FIE construct includes 2.5 kb upstream and 1.3 kb downstream sequences of the FIE coding region in the pCAMBIA1300 binary vector as previously described for generating transgenic plants51. The triple Flag-tag sequence was introduced along with the promoter sequence and GFP was inserted between the Flag sequence and the first ATG codon sequences. The phosphorylation site mutation pFIE-Flag-GFP-FIE (SSTS/AAAA) construct was generated by replacing the FIE sequence using site-directed mutagenesis. These binary plasmids were transformed into heterozygote fie+/− (GK-534F01-020364) mutant plants by using the Agrobacterium tumefaciens (strain GV3101)-mediated floral-dip method52. Positive transformants were selected based on hygromycin resistance and confirmed by immunoblot analysis using an anti-Flag antibody. At the T3 generation, GFP-FIE/fie and SSTS/AAAA/fie plants were confirmed by genotyping using primers listed in Supplementary Table 8.
pFIE:Flag-GFP-FIE 构建体包括 pCAMBIA1300 二元载体中 FIE 编码区的 2.5 kb 上游序列和 1.3 kb 下游序列,如前所述,用于生成转基因植物51。三重 Flag 标签序列与启动子序列一起引入,并在 Flag 序列和第一个 ATG 密码子序列之间插入 GFP。磷酸化位点突变 pFIE-Flag-GFP-FIE (SSTS/AAAA) 构建体是通过定点诱变替换 FIE 序列生成的。通过使用根癌农杆菌 (菌株 GV3101) 介导的 floral-dip 法52,将这些二元质粒转化到杂合子 fie+/- (GK-534F01-020364) 突变植物中。根据潮霉素耐药性选择阳性转化体,并使用抗 Flag 抗体通过免疫印迹分析确认。在 T3 代,通过使用补充表 8 中列出的引物进行基因分型来确认 GFP-FIE/fie 和 SSTS/AAAA/fie 植物。
To generate the oestradiol-inducible FIE artificial miRNA mutant (fie-amiR-es) transgenic plants, optimal amiRNAs were first selected using the epitope-tagged protein-based amiRNA (ETPamir) screens in protoplast assays53. To generate the amiRNA expression constructs, candidate amiRNA sequences were cloned into the pUC119-RCS plasmid. To generate the target construct for amiRNA screens, the coding region of the FIE cDNA was PCR amplified and cloned into the plasmid with a heat shock promoter (pHSP) to generate pHSP-FIE-Flag-NOS. The pHBT-GFP-HA plasmid was used as a control for protoplast co-transfection and internal control as described53. After screening in protoplast, the optimal amiRNA was amplified and transferred to the oestradiol-inducible vector pLB12 (ref. 54). Positive transformants were selected based on kanamycin resistance.
为了生成雌二醇诱导型 FIE 人工 miRNA 突变体 (fie-amiR-es) 转基因植物,首先在原生质体测定中使用表位标记的基于蛋白质的 amiRNA (ETPamir) 筛选选择最佳 amiRNA 53。为了生成 amiRNA 表达构建体,将候选 amiRNA 序列克隆到 pUC119-RCS 质粒中。为了生成用于 amiRNA 筛选的靶构建体,对 FIE cDNA 的编码区进行 PCR 扩增,并使用热休克启动子 (pHSP) 克隆到质粒中,以生成 pHSP-FIE-Flag-NOS。pHBT-GFP-HA 质粒用作原生质体共转染和内部对照的对照,如 53 所述。在原生质体中筛选后,扩增最佳 amiRNA 并将其转移到雌二醇诱导载体 pLB12 上(参考文献 54)。根据卡那霉素抗性选择阳性转化体。
For the PUP-IT proximity-tagging system27 (for Fig. 2a,b), the sequences of p2X35S-3×Flag-pup (E) and pUBQ10-PafA-HA were synthesized and inserted into the pCAMBIA1300 binary vector, resulting in pCambia-PUP-IT. The sequence encoding the C terminus (1226–2480) of TOR (TOR-C) was amplified by PCR to generate the plasmid with pUBQ10-TOR-C-PafA-HA and p2X35S-3×Flag-pup (E). The coding region sequences of PRC2 components, including CLF, SWN, EMF2, VRN2, FIE and MSI1, were amplified by PCR and cloned into the pHBT-MYC plant expression vector55. For the confocal imaging of FIE mutants in mesophyll protoplasts, the coding region of FIE was first amplified by PCR and cloned into the pHBT-GFP expression vector, resulting in pHBT-GFP-FIE. The mutant variants were generated by site-directed mutagenesis using pHBT-GFP-FIE as a template. For the expression of recombinant FIE protein and its mutant variant proteins in Escherichia coli, the coding region of FIE was first amplified by PCR and cloned into the pET14b expression vector, resulting in pET14b-FIE. The mutant variants were generated by site-directed mutagenesis using pET14b-FIE as a template. We produced the recombinant protein for each of four core subunits of the plant PRC2 complexes using the SF9 insect cells. The coding sequences of FIE and its phosphorylation site mutation SSTS/AAAA with 3×Flag tags at the N terminus were cloned into the pFastBac1 vector. The coding sequences of the other PRC2 subunits CLF, EMF2 and MSI1 were cloned into the pFastBac1 vector with His tags at their N terminus. All constructs were verified by Sanger-sequencing. The primers used for plasmid construction and site-directed mutagenesis are listed in Supplementary Table 8.
对于 PUP-IT 接近标记系统27(对于 Fig.2a,b),合成 p2X35S-3×Flag-pup (E) 和 pUBQ10-PafA-HA 的序列并将其插入 pCAMBIA1300 二元载体中,得到 pCambia-PUP-IT。通过 PCR 扩增编码 TOR (TOR-C) 的 C 末端 (1226–2480) 的序列,以生成带有 pUBQ10-TOR-C-PafA-HA 和 p2X35S-3×Flag-pup (E) 的质粒。通过 PCR 扩增 PRC2 组分的编码区序列,包括 CLF 、 SWN 、 EMF2 、 VRN2 、 FIE 和 MSI1 ,并克隆到 pHBT-MYC 植物表达载体中55。对于叶肉原生质体中 FIE 突变体的共聚焦成像,首先通过 PCR 扩增 FIE 的编码区并克隆到 pHBT-GFP 表达载体中,得到 pHBT-GFP-FIE。突变变体是使用 pHBT-GFP-FIE 作为模板通过定点诱变生成的。对于重组 FIE 蛋白及其突变变体蛋白在大肠杆菌中的表达,首先通过 PCR 扩增 FIE 的编码区,并将其克隆到 pET14b 表达载体中,得到 pET14b-FIE。突变变体是使用 pET14b-FIE 作为模板通过定点诱变生成的。我们使用 SF9 昆虫细胞为植物 PRC2 复合物的四个核心亚基中的每一个生产了重组蛋白。将 FIE 的编码序列及其磷酸化位点突变 SSTS/AAAA 在 N 端带有 3×Flag 标签克隆到 pFastBac1 载体中。将其他 PRC2 亚基 CLF 、 EMF2 和 MSI1 的编码序列克隆到 pFastBac1 载体中,其 N 端为 His 标签。 所有构建体均通过 Sanger 测序验证。用于质粒构建和定点诱变的引物列在补充表 8 中。
Plant materials 植物材料
The A. thaliana ecotype Columbia (Col-0) was used as wild-type (WT) plants in this study, unless otherwise stated. The oestradiol-inducible RNAi tor-es mutant and the S6K1-HA transgenic line have been described previously21. The transgenic plants expressing PRC2 components, pSWN:SWN-GFP56, p35S:GFP-CLF57, pEMF2:EMF2-GFP58 and pMSI1:MSI1-GFP58 have been described previously. The T-DNA null mutants in canonical TORC1, including raptor1 (SALK_078159)24, raptor2 (SALK_043920)24, lst8-1-2 (SAIL_641_D10)25 and lst8-2 (SALK_018605), were confirmed by genotyping and RT–qPCR. Transgenic plants GFP-FIE/fie, SSTS/AAAA/fie and fie-amiR-es were generated as described below. Col-FRISF2 (Col-FRI)42 and pFLC:FLC-GUS in Ler-FRI59 lines were used for the vernalization analysis. The pFIE:Flag-GFP-FIE construct was introduced into Col-FRI, tor-es or fie-amiR-es by genetic crossing, antibiotic selected, and confirmed by PCR-based genotyping and immunoblot analyses. Col-FRI was crossed with S6K1-HA, tor-es, GFP-FIE/fie or SSTS/AAAA/fie and confirmed by PCR-based genotyping and immunoblot analyses. The pKNAT1:GUS60 line was introduced into GFP-FIE/fie and SSTS/AAAA/fie by genetic crossing and confirmed by PCR-based genotyping.
除非另有说明,否则 A. thaliana 生态型 Columbia (Col-0) 在本研究中用作野生型 (WT) 植物。雌二醇诱导型 RNAi tor-es 突变体和 S6K1-HA 转基因系之前已经描述过21。表达 PRC2 成分 pSWN:SWN-GFP56、p35S:GFP-CLF57、pEMF2:EMF2-GFP58 和 pMSI1:MSI1-GFP58 的转基因植物之前已经描述过。通过基因分型和 RT-qPCR 证实经典 TORC1 中的 T-DNA 缺失突变体,包括 raptor1 (SALK_078159)24 、 raptor2 (SALK_043920)24 、 lst8-1-2 (SAIL_641_D10)25 和 lst8-2 (SALK_018605)。转基因植物 GFP-FIE/fie、SSTS/AAAA/fie 和 fie-amiR-es 的生成如下所述。使用 Ler-FRI59 系中的 Col-FRISF2 (Col-FRI)42 和 pFLC:FLC-GUS 进行春化分析。通过基因杂交、抗生素选择将 pFIE:Flag-GFP-FIE 构建体引入 Col-FRI、tor-es 或 fie-amiR-es 中,并通过基于 PCR 的基因分型和免疫印迹分析确认。Col-FRI 与 S6K1-HA 、 tor-es 、 GFP-FIE/fie 或 SSTS/AAAA/fie 杂交,并通过基于 PCR 的基因分型和免疫印迹分析进行确认。通过基因杂交将 pKNAT1:GUS60 系引入 GFP-FIE/fie 和 SSTS/AAAA/fie,并通过基于 PCR 的基因分型进行确认。
Plant growth conditions 植物生长条件
Seeds were surface sterilized with 70% ethanol and bleach (25% bleach, 0.02% Triton X-100 for 10 min), washed with sterile water three times, and stratified at 4 °C for three days before plating. Sterilized seeds were planted and germinated in a nutrient rich sugar-containing medium, liquid 0.5× MS medium (0.5× MS, 2 mM MES and 0.5% sucrose, pH 5.7) or on solid 0.5× MS medium (0.5× MS, 2 mM MES, 0.5% sucrose and 0.6% phytoagar, pH 5.7). To analyse the effect of TOR activity on plant growth and development (for Fig. 1a and Extended Data Fig. 1a,b), Torin2 at indicated concentrations (0, 0.1, 0.5, 1, 5 and 10 μM), 10 μM oestradiol, or DMSO only, were added at the time of seed germination for 8 days. To quantify the histone methylation marks and gene expressions in WT and the tor-es mutant (for Fig. 1b,c and Extended Data Figs. 1a–d, 3b and 8d,e), seeds were planted and germinated in liquid 0.5× MS medium (0.5× MS, 2 mM MES, and 0.5% sucrose, pH 5.7) for 7 days. On the fourth day, 10 μM oestradiol was added to the medium to deplete TOR in tor-es plants. For the sugar starvation experiment (for Fig. 1d,e and Extended Data Figs. 2e and 8f,g), WT, S6K1-HA or the indicated mutant Arabidopsis seedlings were grown in sugar-free liquid 0.5× MS medium (0.5× MS, 2 mM MES, pH 5.7) for 7 days. The endogenous glucose from the seed is depleted three days after germination4. Glucose (25 mM) was added on the eighth day for the indicated time (for Fig. 1d,e) or 6 h (for Extended Data Figs. 2e and 8f,g) to stimulate TOR signalling. For the experiments with TOR or S6K inhibitors (for Fig. 1f,g and Extended Data Fig. 2a–d), WT or S6K1-HA seedlings were grown in liquid 0.5× MS (with 0.5% sucrose, PH 5.7) medium for four days and treated with DMSO as the control or 10 μM of different chemical inhibitors that target TOR or S6K1 for three days. Unless otherwise indicated, plants were grown at 23/20 °C, 12 h/12 h light/dark, 60% relative humidity and 75 μmol m−2 s−1 light.
用 70% 乙醇和漂白剂(25% 漂白剂,0.02% Triton X-100 10 10 分钟)对种子进行表面灭菌,用无菌水洗涤 3 次,并在 4 °C 下分层 3 天,然后铺板。将灭菌的种子种植在营养丰富的含糖培养基、液体 0.5× MS 培养基(0.5× MS、2 mM MES 和 0.5% 蔗糖,pH 5.7)或 0.5× MS 固体培养基(0.5× MS、2 mM MES、0.5% 蔗糖和 0.6% 植物琼脂,pH 5.7)中发芽。为了分析 TOR 活性对植物生长和发育的影响(图 D)。1a 和扩展数据图 1.1a,b)、在种子发芽时添加指定浓度(0、0.1、0.5、1、5 和 10 μM)的 Torin2、10 μM 雌二醇或仅 DMSO,持续 8 天。为了量化 WT 和 tor-es 突变体中的组蛋白甲基化标记和基因表达(对于 Fig.1b,c 和扩展数据图1a–d、3b 和 8d,e),将种子种植并在液体 0.5× MS 培养基(0.5× MS、2 mM MES 和 0.5% 蔗糖,pH 5.7)中发芽 7 天。第四天,向培养基中加入 10 μM 雌二醇以消耗 tor-es 植物中的 TOR。对于糖饥饿实验(图 D)。1d,e 和扩展数据图2e 和 8f,g)、WT、S6K1-HA 或指定的突变拟南芥幼苗在无糖液体 0.5× MS 培养基(0.5× MS,2 mM MES,pH 5.7)中生长 7 天。种子中的内源性葡萄糖在发芽后 3 天耗尽4。在第 8 天添加葡萄糖 (25 mM) 指定时间(对于 Fig.1d,e)或 6 小时(对于扩展数据图2e 和 8f,g) 刺激 TOR 信号传导。对于 TOR 或 S6K 抑制剂的实验(对于 Fig.1f,g 和扩展数据图 2a-d),WT 或 S6K1-HA 幼苗在 0.5× MS (含 0.5% 蔗糖,PH 5.7) 液体培养基中生长 4 天,并用 DMSO 作为对照或 10 μM 靶向 TOR 或 S6K1 的不同化学抑制剂处理 3 天。除非另有说明,否则植物在 23/20 °C、12 h/12 h 光照/黑暗、60% 相对湿度和 75 μmol m -2 s -1 光照下生长。
External data sources 外部数据源
The sequences of proteins were obtained from TAIR (https://www.arabidopsis.org/) and NCBI (https://www.ncbi.nlm.nih.gov/). The 3D protein structures were predicted by RaptorX (http://raptorx.uchicago.edu//).
蛋白质序列从 TAIR (https://www.arabidopsis.org/) 和 NCBI (https://www.ncbi.nlm.nih.gov/) 获得。3D 蛋白质结构由 RaptorX (http://raptorx.uchicago.edu//) 预测。
Generation, purification and application of the pFIE(S14) antibody
pFIE(S14) 抗体的生成、纯化和应用
The phospho-site-specific anti-pFIE(S14) antibody was custom-made by ABclonal. In brief, a phosphopeptide (NESIVGpSLTPSN-C) was synthesized and conjugated to keyhole limpet haemocyanin carrier for the immunization of four rabbits. The polyclonal antiserum was first affinity-purified using the phosphopeptide, and the elution was then passed over the column coupled with the non-phospho-peptide (NESIVGSLTPSN-C) to remove nonspecific antibodies61. The pFIE(S14) antibody was first tested by dot blot assays against the phospho- and non-phospho-peptides. The anti-pFIE(S14) antibody was further validated by immunoblot analyses against the His–FIE or His–FIE(S14A) proteins without or with TOR phosphorylation after the in vitro kinase assay.
磷酸位点特异性抗 pFIE(S14) 抗体由 ABclonal 定制。简而言之,合成了磷酸肽 (NESIVGpSLTPSN-C) 并与锁孔帽贝血蓝蛋白载体偶联,用于免疫 4 只兔子。首先使用磷酸肽对多克隆抗血清进行亲和纯化,然后将洗脱液与非磷酸肽 (NESIVGSLTPSN-C) 偶联通过色谱柱以去除非特异性抗体61。pFIE(S14) 抗体首先通过斑点印迹法检测磷酸肽和非磷酸肽。在体外激酶测定后,通过针对 His-FIE 或 His-FIE(S14A) 蛋白的免疫印迹分析进一步验证了抗 pFIE(S14) 抗体,而没有或有 TOR 磷酸化。
To detect the phosphorylated pFIE(S14) in vivo, pFIE(S14) antibody was used to detect the Flag–GFP–FIE protein immunoprecipitated from transgenic plants by immunoblot analyses. Specifically, GFP-FIE/fie or tor-es seedlings expressing Flag–GFP–FIE were grown in liquid 0.5× MS (with 0.5% sucrose, PH 5.7) medium for eight days (for Fig. 2g). The medium was changed to liquid 0.5× MS without sugar and the seedlings were starved in dark for two days. Glucose (25 mM) was added to the medium for 2 h to stimulate TOR activity. To deplete TOR, tor-es seedlings was induced by 10 μM β-oestradiol for four days. For Torin2 treatment, 2 μM Torin2 was added into the medium one day before collecting the samples. To examine glucose-induced FIE phosphorylation (for Fig. 2h and Extended Data Fig. 5d), GFP-FIE/fie seedlings were grown in sugar-free liquid 0.5× MS medium (0.5× MS, 2 mM MES, pH 5.7) for 7 days. Glucose (25 mM) was added to the medium for 2 h on the 8th day to stimulate TOR signalling.
为了检测体内磷酸化的 pFIE(S14),使用 pFIE(S14) 抗体通过免疫印迹分析检测从转基因植物免疫沉淀的 Flag-GFP-FIE 蛋白。具体来说,表达 Flag-GFP-FIE 的 GFP-FIE/fie 或 tor-es 幼苗在 0.5× MS 液体(含 0.5% 蔗糖,PH 5.7)培养基中生长 8 天(图 D)。将培养基改为无糖的 0.5× MS 液体,将幼苗避光两天。将葡萄糖 (25 mM) 加入培养基中 2 小时以刺激 TOR 活性。为了消耗 TOR,用 10 μM β-雌二醇诱导 tor-es 幼苗 4 天。对于 Torin2 处理,在收集样品前一天向培养基中加入 2 μM Torin2。为了检查葡萄糖诱导的 FIE 磷酸化(图 D)。2h 和扩展数据图5d),将 GFP-FIE/fie 幼苗在 0.5× MS 培养基 (0.5× MS, 2 mM MES, pH 5.7) 的无糖液体中培养 7 天。第 8 天将葡萄糖 (25 mM) 加入培养基中 2 小时以刺激 TOR 信号传导。
Approximately 5 g (about 1,000) seedlings were collected and ground to fine powder with a mortar and pestle in liquid nitrogen and suspended in 5 ml of extraction buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 5 mM EDTA, 10 mM p-nitrophenyl phosphate, 20 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 2 mM Na3VO4, 1 mM NaF, 1% NP-40, 10% glycerol, 1×protease inhibitor cocktail tablet, and 1× Phosphatase Inhibitor Cocktail Tablet (PhosSTOP)). The tissues were further homogenized by grinding thoroughly and then centrifuged in an SS34 rotor for 25 min at 12,500 rpm and the supernatant was saved. The pellets were resuspended in 2 ml lysis buffer (25 mM Tris-HCl, pH 7.6, 75 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 10 mM p-nitrophenyl phosphate, 20 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 2 mM Na3VO4, 1 mM NaF, 10% glycerol, 1× protease inhibitor cocktail tablet, and 1× Phosphatase Inhibitor Cocktail Tablet (PhosSTOP)) and sonicated for 1 min (10s on, 20s off). The supernatant from the two-step extraction was combined and incubated with 100 ml of pre-washed anti-Flag M2 Agarose Beads (Sigma, A2220) at 4 °C for 2 h with rotation. Beads were washed five times with extraction buffer. Proteins were then eluted from the anti-Flag M2 Agarose beads with 50 μl of 300 mg ml−1 of 3×Flag peptide (Sigma, F 4799) five times at room temperature. The elution was precipitated with an equal volume of 20% TCA/acetone and washed with acetone. The pellet was resuspended in 100 μl of 1× calf intestinal alkaline phosphatase (CIP) buffer. For CIP treatment, 50 μl of Flag–GFP–FIE protein solution was mixed with or without (mock) 10 U CIP (New England Biolabs, M0290) for 1 h at 37 °C. The reaction was stopped by adding 2× sample buffer and boiled for 5 min at 95 °C before running the SDS–PAGE. Site-specific phosphorylation was detected by immunoblot analysis with pFIE(S14) antibody at 1:500 dilution in 5% BSA.
收集约 5 g(约 1,000 株)幼苗,用研钵和研杵在液氮中研磨成细粉,并悬浮于 5 ml 提取缓冲液(25 mM Tris-HCl pH 7.6、150 mM NaCl、5 mM EDTA、10 mM 对硝基苯磷酸酯、20 mM β-甘油磷酸盐、10 mM 焦磷酸钠、2 mM Na3VO 4 、 1 mM NaF、1% NP-40、10% 甘油、1×蛋白酶抑制剂混合物片剂和 1× 磷酸酶抑制剂混合物片剂 (PhosSTOP))。通过彻底研磨将组织进一步匀浆,然后在 SS34 转子中以 12,500 rpm 离心 25 分钟,并保存上清液。将沉淀重悬于 2 ml 裂解缓冲液(25 mM Tris-HCl,pH 7.6、75 mM NaCl、5 mM EDTA、1% Triton X-100、0.1% SDS、10 mM 对硝基苯磷酸酯、20 mM β-甘油磷酸酯、10 mM 焦磷酸钠、2 mM Na3VO 4 、1 mM NaF、10% 甘油、1×蛋白酶抑制剂混合物片剂和 1× 磷酸酶抑制剂混合物片剂 (PhosSTOP)) 中并超声处理 1 分钟(10 秒后, 20 秒)。合并来自两步提取的上清液,并与 100 ml 预洗的抗 Flag M2 琼脂糖珠(Sigma,A2220)在 4 °C 下旋转孵育 2 小时。用提取缓冲液洗涤珠子 5 次。然后在室温下用 50 μl 300 mg ml-1 的 3×Flag 肽 (Sigma, F 4799) 从抗 Flag M2 琼脂糖珠中洗脱蛋白质 5 次。用等体积的 20% TCA/丙酮沉淀洗脱液,并用丙酮洗涤。将沉淀重悬于 100 μl 1× 小牛肠碱性磷酸酶 (CIP) 缓冲液中。对于 CIP 处理,将 50 μl Flag-GFP-FIE 蛋白溶液与或不与(模拟)10 U CIP(New England Biolabs,M0290)在 37 °C 下混合 1 小时。 通过加入 2× 样品缓冲液终止反应,并在 95 °C 下煮沸 5 分钟,然后运行 SDS-PAGE。用 pFIE(S14) 抗体在 5% BSA 中以 1:500 稀释度进行免疫印迹分析,检测位点特异性磷酸化。
RNA extraction and gene expression analysis
RNA 提取和基因表达分析
Whole seedlings from liquid medium or the aerial parts of plants from the agar plate were collected and frozen in liquid nitrogen. Total RNA was isolated from seedlings ground in liquid nitrogen and extracted using the protocol with the Trizol reagent (Invitrogen). Total RNA (0.5 ug/20 ul reaction) was treated with DNase I (RQ1 RNase-free DNase I, Promega), and converted to cDNA using M-MLV reverse transcriptase (RNase H minus, Point Mutant, Promega) and oligo(dT) primer according to the manufacturer’s guidelines. Quantitative PCR was carried out as described62 using the primers listed in Supplementary Table 8. The relative gene expression was normalized to the expression of UBQ10 (AT4G05320), UBC21 (At5g25760) or ACTIN2 (At5g09810). A minimum of triplicate biological samples were analysed with consistent results.
收集来自液体培养基的整株幼苗或琼脂平板中植物的地上部分,并在液氮中冷冻。从液氮研磨的幼苗中分离总 RNA,并使用 Trizol 试剂 (Invitrogen) 的方案提取。用 DNase I (RQ1 RNase free DNase I, Promega) 处理总 RNA (0.5 ug/20 ul 反应),并使用 M-MLV 逆转录酶 (RNase H minus, Point Mutant, Promega) 和 oligo (dT) 引物转化为 cDNA根据制造商的指南。使用补充表 8 中列出的引物按照62 所述进行定量 PCR。将相对基因表达标准化为 UBQ10 (AT4G05320) 、 UBC21 (At5g25760) 或 ACTIN2 (At5g09810) 的表达。分析了最少一式三份的生物样品,结果一致。
To analyse the perturbed global gene expression pattern in the fie-amiR-es and SSTS/AAAA/fie mutants, RNA-seq analyses were carried out as described51. Seedlings of WT, fie-amiR-es, GFP-FIE/fie or STSS/AAAA/fie were grown on 0.5× MS (with 0.5% sucrose, pH 5.7) agar medium for two weeks (For Fig. 3a–e,g and Extended Data Fig. 7a–c). For the inducible fie-amiR-es mutant, 10 mM oestradiol was added into the medium at the beginning of germination to induce FIE depletion. The aerial part of plants was collected for RNA extraction at two weeks. Total RNA (0.5 μg) was used for preparing the library with NEBNext Ultra II RNA Library Prep Kit for Illumina sequencing according to the manufacturer’s guidelines. For the DNA fragment enrichment step, the template was amplified by PCR for eight cycles with adaptors of different barcodes. The libraries were sequenced using an Illumina HiSeq4000 at GENEWIZ Next Generation Sequencing centre, and 150 bp paired-end reads were generated.
为了分析 fie-amiR-es 和 SSTS/AAAA/fie 突变体中受干扰的全局基因表达模式,按所述进行 RNA-seq 分析51。WT、fie-amiR-es、GFP-FIE/fie 或 STSS/AAAA/fie 的幼苗在 0.5× MS(含 0.5% 蔗糖,pH 5.7)琼脂培养基上生长两周(图 D)。3a–e,g 和扩展数据图7a-c)。对于可诱导型 fie-amiR-es 突变体,在发芽开始时向培养基中加入 10 mM 雌二醇以诱导 FIE 耗竭。在两周时收集植物的地上部分用于 RNA 提取。根据制造商的指南,使用总 RNA (0.5 μg) 和 NEBNext Ultra II RNA 文库制备试剂盒制备文库,用于 Illumina 测序。对于 DNA 片段富集步骤,使用不同条形码的接头通过 PCR 扩增模板 8 个循环。在 GENEWIZ Next Generation Sequencing 中心使用 Illumina HiSeq4000 对文库进行测序,并生成 150 bp 的双端读长。
ChIP analysis ChIP 分析
ChIP–seq data were processed and analysed as previously described with minor modification63. In brief, plants (1 g) were ground into fine powder in liquid nitrogen and suspended with 10 ml nuclei isolation buffer (10 mM HEPES pH 8.0, 1 M sucrose, 5 mM KCl, 5 mM EDTA, 0.6% Triton X-100, 0.4 mM PMSF, and fresh protease inhibitor cocktail). The homogenate was incubated with 1% formaldehyde for 15 min and stopped with 125 mM glycine. The extract was filtered through double layers of wet Miracloth and centrifuged at 1,500 g for 10 min at 4 °C. The nuclei pellet was washed 3 times with 10 ml nuclei isolation buffer. The pellet was resuspended in 0.2 ml nuclei lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS, and fresh protease inhibitor cocktail) and sonicated for 3 × 10 min, 30s on 30s off at high setting, and then diluted 10-fold to 2 ml with ChIP dilution buffer (1.1 % Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.0, 167 mM NaCl, and fresh protease inhibitor cocktail). Chromatin was immunoprecipitated with an antibody at 4 °C overnight with rotation. Each antibody, against H3 (Abcam, 1791,1:1,000), H3K27me3 (Millipore, 07-449,1:200) and H3K9me2 (Abcam 1220, 1:200), was bound to the pre-washed protein A or G Dyna-beads (40 μl) for 1 h at 4 °C with rotation. The immunoprecipitated chromatin was washed twice with each of the following solutions for 5 min at 4 °C: 1 ml of low-salt buffer, high-salt buffer, LiCl buffer and TE buffer. Protein complexes were eluted from beads with 400 µl of elution buffer (1% SDS and 0.1 M NaHCO3) at 65 °C for 10 min. The eluted chromatin was reverse crosslinked with 0.2 M NaCl at 65 °C overnight, and treated with RNase A (0.2 mg ml−1) at 37 °C for 1 h, proteinase K at 50 °C for 1 h, and purified with phenol:chlorophorm followed by ethanol precipitation. The enriched DNA was subjected to quantitative PCR (qPCR) analysis or library construction and Illumina sequencing for ChIP–seq analysis.
如前所述处理和分析 ChIP-seq 数据,并稍作修改63。简而言之,将植物 (1 g) 在液氮中研磨成细粉,并用 10 ml 细胞核分离缓冲液(10 mM HEPES pH 8.0、1 M 蔗糖、5 mM KCl、5 mM EDTA、0.6% Triton X-100、0.4 mM PMSF 和新鲜蛋白酶抑制剂混合物)悬浮。将匀浆与 1% 甲醛孵育 15 分钟,然后用 125 mM 甘氨酸终止。通过双层湿 Miracloth 过滤提取物,并在 4°C 下以 1,500 g 离心 10 分钟。 用 10 ml 细胞核分离缓冲液洗涤细胞核沉淀 3 次。将沉淀重悬于 0.2 ml 细胞核裂解缓冲液(50 mM Tris-HCl pH 8.0、10 mM EDTA、1% SDS 和新鲜蛋白酶抑制剂混合物)中,超声处理 3 × 10 分钟,在高设置下关闭 30 秒,然后用 ChIP 稀释缓冲液(1.1% Triton X-100、1.2 mM EDTA、 16.7 mM Tris-HCl pH 8.0、167 mM NaCl 和新鲜蛋白酶抑制剂混合物)。用抗体在 4 °C 下旋转染色质免疫沉淀过夜。针对 H3 (Abcam, 1791,1:1,000)、H3K27me3 (Millipore, 07-449,1:200) 和 H3K9me2 (Abcam 1220, 1:200) 的每种抗体在 4 °C 下与预洗的蛋白 A 或 G Dyna 珠 (40 μl) 结合 1 小时,并旋转。免疫沉淀的染色质在 4 °C 下用以下每种溶液洗涤两次,每次 5 分钟:1 ml 低盐缓冲液、高盐缓冲液、LiCl 缓冲液和 TE 缓冲液。在 65 °C 下用 400 μl 洗脱缓冲液(1% SDS 和 0.1 M NaHCO3)从珠子中洗脱蛋白质复合物 10 分钟。 洗脱的染色质在 65 °C 下用 0.2 M NaCl 反向交联过夜,并在 37 °C 下用 RNase A (0.2 mg ml-1) 处理 1 小时,用蛋白酶 K 在 50 °C 下处理 1 小时,并用苯酚:氯酚纯化,然后乙醇沉淀。对富集的 DNA 进行定量 PCR (qPCR) 分析或文库构建和 Illumina 测序以进行 ChIP-seq 分析。
ChIP–qPCR was performed using the iQ SYBR green supermix (Bio-Rad) and normalized using H3 level as an internal standard. H3K27me3 enrichment is shown as the percentage of H3. Three biological replicates were performed for each experiment. Primer sequences used for ChIP–qPCR are listed in Supplementary Table 8.
使用 iQ SYBR 绿色超混合液 (Bio-Rad) 进行 ChIP-qPCR,并使用 H3 水平作为内标进行归一化。H3K27me3 富集显示为 H3 的百分比。每个实验进行 3 次生物学重复。用于 ChIP-qPCR 的引物序列列于补充表 8 中。
To quantitatively compare genome-wide H3K27me3 levels across plant samples, a method called ChIP with reference exogenous genome (ChIP-Rx) was performed with minor modifications as previously described23. Defined quantities (one-tenth) of the reference chromatin from Human 293T cells were added into Arabidopsis chromatin from 1 g of seedlings before immunoprecipitation.
为了定量比较植物样品中的全基因组 H3K27me3 水平,我们进行了一种称为 ChIP 与参考外源基因组 (ChIP-Rx) 的方法,如前所述进行了微小的修改23。在免疫沉淀之前,将来自人 293T 细胞的确定量(十分之一)的参考染色质添加到 1 g 幼苗的拟南芥染色质中。
To determinate the concentration of chromatin, 10 μl from 2 ml of Arabidopsis chromatin was reverse crosslinked, digested with RNase A and proteinase K, and followed by DNA purification as described above. The concentration of purified DNA was determined using Qubit dsDNA High-Sensitivity Assays (Invitrogen). The DNA concentration of 10 μl chromatins from 106 human HEK293T cells was also isolated and determined with the same method. One-tenth of human chromatin (about 10 µg DNA) compared with the Arabidopsis chromatin was mixed with each of different Arabidopsis samples (about 100 µg DNA) before immunoprecipitation. Purified DNA (5 ng) after ChIP was used for preparing the library with NEBNext Ultra II DNA Library Prep Kit for Illumina according to the manufacturer’s guidelines. For the DNA fragments enrichment step, the templates were amplified for eight cycles with adaptors of different barcodes. The libraries were sequenced using an Illumina HiSeq4000 at GENEWIZ Next Generation Sequencing centre and 150 bp pair-end reads were generated.
为了测定染色质的浓度,从 2 ml 拟南芥染色质中取 10 μl,用 RNase A 和蛋白酶 K 消化,然后如上所述进行 DNA 纯化。使用 Qubit dsDNA High-Sensitivity Assays (Invitrogen) 测定纯化 DNA 的浓度。还从 106 个人HEK293T细胞中分离出 10 μl 染色质的 DNA 浓度,并用相同的方法测定。在免疫沉淀之前,将与拟南芥染色质相比的十分之一的人染色质(约 10 μg DNA)与每个不同的拟南芥样品(约 100 μg DNA)混合。根据制造商的指南,使用 ChIP 后纯化的 DNA (5 ng) 与 NEBNext Ultra II DNA 文库制备试剂盒(用于 Illumina)制备文库。对于 DNA 片段富集步骤,使用不同条形码的接头对模板进行 8 个循环扩增。在 GENEWIZ Next Generation Sequencing 中心使用 Illumina HiSeq4000 对文库进行测序,并生成 150 bp 的双端读长。
In vivo co-immunoprecipitation assay
体内免疫共沉淀测定
To capture the dynamic interaction between TOR kinase and substrates in vivo, TOR activity was synchronized and stimulated by starvation and glucose depletion, and formaldehyde crosslinking was applied to stabilize the TOR-substrate interaction. Specifically, transgenic Arabidopsis seedlings expressing Flag–GFP–FIE or Col-0 seedlings as the control were grown in liquid 0.5× MS (with 0.5% sucrose, PH 5.7) medium for eight days (for Fig. 2c). Then the liquid medium was changed to 0.5× MS without sugar and the seedlings were starved in dark for 3 days. Glucose (25 mM) was added into the medium for 2 h to stimulate TOR activity. To test the effect of glucose concentration on the interaction between TOR and FIE (Extended Data Fig. 4a), transgenic seedlings expressing Flag–GFP–FIE were grown in sugar-free liquid 0.5× MS medium (0.5× MS, 2 mM MES, pH 5.7) for 7 days. Different concentration of glucose (0, 0.1, 0.5, 1, 5, 10, 20 and 25 mM) were added to the medium for 2 h on the eighth day to stimulate TOR signalling. Whole seedlings were crosslinked in 1% formaldehyde for 15 min and collected after removing all the liquid with Kimwipes. Crosslinked plants were ground into powder with liquid nitrogen and suspended in extraction buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 0.5 mM DTT, and fresh protease inhibitor cocktail). After centrifugation at 18,000 rpm in a microfuge at 4 °C for 20 min, the supernatant was incubated with pre-washed anti-Flag M2 Agarose Beads (Sigma, A2220) at 4 °C for 2 h with rotation. Beads were washed five times with extraction buffer. Protein complexes were eluted twice with 3×Flag peptide. The eluted and input samples were mixed with 2X sample buffer and boiled for 10 min at 95 °C before SDS–PAGE and immunoblot analysis.
为了捕获体内 TOR 激酶与底物之间的动态相互作用,通过饥饿和葡萄糖耗竭来同步和刺激 TOR 活性,并应用甲醛交联来稳定 TOR-底物相互作用。具体来说,表达 Flag-GFP-FIE 或 Col-0 幼苗作为对照的转基因拟南芥幼苗在 0.5× MS(含 0.5% 蔗糖,PH 5.7)培养基中生长 8 天(对于 Fig.然后将液体培养基改为 0.5× MS 无糖,幼苗避光 3 天。将葡萄糖 (25 mM) 加入培养基中 2 小时以刺激 TOR 活性。为了测试葡萄糖浓度对 TOR 和 FIE 之间相互作用的影响(扩展数据图 1)。4a),表达 Flag-GFP-FIE 的转基因幼苗在 0.5× MS 培养基(0.5× MS,2 mM MES,pH 5.7)的无糖液体中生长 7 天。在第 8 天向培养基中加入不同浓度的葡萄糖 (0 、 0.1 、 0.5 、 1 、 5 、 10 、 20 和 25 mM) 2 小时以刺激 TOR 信号传导。将整株幼苗在 1% 甲醛中交联 15 分钟,并在用 Kimwipes 除去所有液体后收集。将交联植物用液氮研磨成粉末,并悬浮于提取缓冲液(25 mM Tris-HCl pH 7.6、150 mM NaCl、5 mM MgCl2、10% 甘油、0.1% NP-40、0.5 mM DTT 和新鲜蛋白酶抑制剂混合物)中。在微量离心机中于4°C下以18,000rpm离心20分钟后,将上清液与预洗的抗Flag M2琼脂糖珠(Sigma,A2220)在4°C下旋转孵育2小时。用提取缓冲液洗涤珠子 5 次。用 3×Flag 肽洗脱蛋白质复合物两次。 将洗脱的样品和输入样品与 2X 样品缓冲液混合,并在 95 °C 下煮沸 10 分钟,然后进行 SDS-PAGE 和免疫印迹分析。
Protein expression and purification
蛋白表达和纯化
Recombinant proteins were expressed in E. coli Rosetta 2 (DE3) pLysS Cells (Novagen). E. coli cells transfected with expression plasmids were first grown in 80% LB/20% TB medium with ampicillin (100 µg ml−1) at 37 °C until reaching 0.6 OD600. The cell culture was cooled to 18 °C and protein expression was induced with 0.5 mM isopropyl β-d-1- thiogalactopyranoside for 18 h. Cells were collected, resuspended in lysis buffer (50 mM pH 7.4 HEPES, 300 mM NaCl, 20 mM imidazole, and fresh protease inhibitor cocktail), and lysed by sonication. The supernatants were incubated with Ni-NTA agarose (Qiagen) for 2 h at 4 °C. The Ni-NTA agarose was washed four times with washing buffer (50 mM pH 7.4 HEPES, 300 mM NaCl, 40 mM imidazole). The recombinant protein was eluted with elution buffer (50 mM pH 7.4 HEPES, 300 mM NaCl, 250 mM imidazole, and fresh protease inhibitor cocktail) for 10 min at room temperature and concentrated by Amicon Ultra-15 10K filter (Millipore). Proteins were stored at −80 °C after being flash-frozen in liquid nitrogen.
重组蛋白在大肠杆菌 Rosetta 2 (DE3) pLysS 细胞 (Novagen) 中表达。首先在 37 °C 下,在含氨苄青霉素 (100 μg ml-1) 的 80% LB/20% TB 培养基中生长转染表达质粒的大肠杆菌细胞,直至达到 0.6 OD600。将细胞培养物冷却至 18 °C,用 0.5 mM 异丙基 β-d-1- 硫代吡喃半乳糖苷诱导蛋白表达 18 h。收集细胞,重悬于裂解缓冲液(50 mM pH 7.4 HEPES、300 mM NaCl、20 mM 咪唑和新鲜蛋白酶抑制剂混合物)中,并通过超声处理裂解。将上清液与 Ni-NTA 琼脂糖 (Qiagen) 在 4 °C 下孵育 2 小时。 用洗涤缓冲液(50 mM pH 7.4 HEPES、300 mM NaCl、40 mM 咪唑)洗涤 Ni-NTA 琼脂糖四次。在室温下用洗脱缓冲液(50 mM pH 7.4 HEPES、300 mM NaCl、250 mM 咪唑和新鲜蛋白酶抑制剂混合物)洗脱重组蛋白 10 分钟,并通过 Amicon Ultra-15 10K 过滤器 (Millipore) 浓缩。蛋白质在液氮中快速冷冻后,在 −80 °C 下储存。
Histone methyltransferase assays with purified PRC2 complexes
使用纯化的 PRC2 复合物进行组蛋白甲基转移酶检测
Each of the four core components of the Arabidopsis PRC2 complexes, including CLF, EMF2, MSI1, and FIE (WT or SSTS/AAAA mutant) were expressed in insect Sf9 cells using the standard Bac-to-Bac baculovirus expression system (Invitrogen). The Sf9 cells were infected with equal amounts of baculovirus for expressing each subunit at the density of 1.8 × 106 cells per ml. The cells were collected and frozen with liquid nitrogen after 72 h incubation at 130 rpm at 27 °C. Insect cells were resuspended in lysis buffer (20 mM HEPES at pH 7.9, 300 mM KCl, 1.5 mM MgCl2, 1 mM PMSF, 0.5 mM DTT, and 0.1% Triton X-100), and sonication 10 × 20 s on ice. After centrifugation at 18,000 rpm in a microfuge at 4 °C for 20 min, the supernatant was incubated with pre-washed anti-Flag M2 Agarose Beads (Sigma, A2220) at 4 °C for 2 h with rotation. Beads were washed five times with extraction buffer. Protein complexes were eluted twice with 3×Flag peptide at room temperature. The protein buffer was changed to storage buffer (20 mM HEPES at pH 7.9, 300 mM KCl, 1.5 mM MgCl2, 10% glycerol) and concentrated by Amicon Ultra-15 10K filter (Millipore). Proteins were stored at −80 °C after being flash-frozen in liquid nitrogen. The Histone lysine methyltransferase assay was performed as previously described64. In brief, 0.3 μg of recombinant PRC2 complexes (with WT or SSTS/AAAA mutant form of FIE) were incubated with 1 μg of H3 (New England Biolabs, M2503S) in the reaction buffer (20 mM HEPES at pH 7.9, 2 mM MgCl2, 1 mM DTT, and 10 μM SAM) for 2 h at 30 °C. The reaction was stopped by addition of 4× loading dye and heated for 5 min at 95 °C. The quantitative level of H3K27me3 was detected by immunoblot analyses using specific anti-H3K27me3 antibody as described above.
拟南芥 PRC2 复合物的四个核心组分,包括 CLF 、 EMF2 、 MSI1 和 FIE (WT 或 SSTS/AAAA 突变体) 使用标准 Bac-to-Bac 杆状病毒表达系统 (Invitrogen) 在昆虫 Sf9 细胞中表达。用等量的杆状病毒感染 Sf9 细胞,以 1.8 × 106 个细胞/ml 的密度表达每个亚基。收集细胞并在 27 °C 下以 130 rpm 孵育 72 小时后用液氮冷冻。 将昆虫细胞重悬于裂解缓冲液(pH 7.9 的 20 mM HEPES、300 mM KCl、1.5 mM MgCl2、1 mM PMSF、0.5 mM DTT 和 0.1% Triton X-100)中,并在冰上超声处理 10 × 20 秒。在微量离心机中于4°C下以18,000rpm离心20分钟后,将上清液与预洗的抗Flag M2琼脂糖珠(Sigma,A2220)在4°C下旋转孵育2小时。用提取缓冲液洗涤珠子 5 次。在室温下用 3×Flag 肽洗脱蛋白质复合物两次。将蛋白质缓冲液改为储存缓冲液(pH 7.9 的 20 mM HEPES、300 mM KCl、1.5 mM MgCl2、10% 甘油)并通过 Amicon Ultra-15 10K 过滤器 (Millipore) 浓缩。蛋白质在液氮中快速冷冻后,在 −80 °C 下储存。组蛋白赖氨酸甲基转移酶测定如前所述进行 64。简而言之,将 0.3 μg 重组 PRC2 复合物(具有 WT 或 SSTS/AAAA 突变形式的 FIE)与 1 μg H3(New England Biolabs,M2503S)在反应缓冲液(pH 值为 7.9 的 20 mM HEPES、2 mM MgCl2、1 mM DTT 和 10 μM SAM)中在 30 °C 下孵育 2 小时。 通过加入 4× 负载染料终止反应,并在 95 °C 下加热 5 分钟。 如上所述,使用特异性抗 H3K27me3 抗体通过免疫印迹分析检测 H3K27me3 的定量水平。
TOR kinase assay TOR 激酶测定
In vitro TOR kinase assay was performed as previously described4. To immunoprecipitate the endogenous TOR complexes, WT seeds were germinated and grown in liquid medium (0.5× MS, 0.5% sucrose, pH 5.7, 6 seedlings in 1 ml in each well in 6-well plates) for 7 days. The same fresh medium (1 ml) was replenished for 2 more h to maximize TOR activity. TOR complexes purified from six seedlings could be used for ten kinase reactions. For in vitro kinase assay (for Fig. 2d,f and Extended Data Fig. 4h), 1 μg of His–FIE or the mutant proteins were incubated with the immunoprecipitated TOR kinase on the beads in the kinase buffer (25 mM HEPES, pH 7.4, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 10 μM cold ATP, 2 μCi [γ-32P]-ATP) at 30 °C for 30 min with shaking at 1,000 rpm. For TOR inhibitor treatment, the immunoprecipitated TOR kinase on the beads was pre-incubated with 1 μM Torin2 for 10 min at room temperature before the kinase assay. The reaction was stopped by adding 2× SDS–PAGE loading buffer. After SDS–PAGE and subsequent gel drying, protein phosphorylation was visualized by autoradiography using the Typhoon imaging system (GE Healthcare).
如前所述进行体外 TOR 激酶测定4.为了免疫沉淀内源性 TOR 复合物,将 WT 种子发芽并在液体培养基 (0.5× MS、0.5% 蔗糖、pH 5.7、6 个幼苗在 6 孔板中每个孔中 1 ml 中)中生长 7 天。将相同的新鲜培养基 (1 ml) 再补充 2 小时以最大限度地提高 TOR 活性。从 6 株幼苗中纯化的 TOR 复合物可用于 10 种激酶反应。对于体外激酶测定(图 D)。2d,f 和扩展数据图4 小时),将 1 μg His-FIE 或突变蛋白与激酶缓冲液(25 mM HEPES,pH 7.4、50 mM KCl、10 mM MgCl2、1 mM DTT、10 μM 冷 ATP、2 μCi [γ-32P]-ATP)中的珠子上的免疫沉淀 TOR 激酶在 30 °C 下孵育 30 分钟,同时以 1,000 rpm 振荡。对于 TOR 抑制剂处理,在激酶测定之前,将珠子上免疫沉淀的 TOR 激酶与 1 μM Torin2 在室温下预孵育 10 分钟。通过加入 2× SDS-PAGE 上样缓冲液终止反应。在 SDS-PAGE 和随后的凝胶干燥后,使用 Typhoon 成像系统 (GE Healthcare) 通过放射自显影显影观察蛋白质磷酸化。
Tandem mass spectrometry analysis
串联质谱分析
To identify the TOR phosphorylation sites in FIE, 5 μg of His–FIE recombinant proteins were subjected to in vitro TOR kinase assay in the kinase buffer for 2 h at 30 °C. The phosphorylated His–FIE was separated by 10% SDS–PAGE gel. The gel was stained with Thermo GelCode Blue Safe Protein Stain and destained with ddH2O. The His–FIE band was excised and subjected to in-gel digestion with trypsin. The phospho-peptides were enriched using the TiO2/ZrO2 media from Glygen65 and subjected to LC–MS/MS analysis using the Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific)66. The raw data of LC–MS/MS spectra was analysed by the Mascot search engine (version 2.2, Matrix Science). The parameters were: Arabidopsis TAIR10 database (32,785 entries, https://www.arabidopsis.org/), precursor mass tolerance at 10 ppm, fragment mass tolerance at 0.8 Da, trypsin with one missed cleavage, oxidation (M) and phosphorylation (S, T, Y) as dynamic modifications. The identified phosphorylated peptides and each phosphorylation site assignment were confirmed by manual inspection of the precursor mass and fragmentation ions. To validate the FIE phosphorylation sites from in vivo immunoprecipitation using Flag antibody, the Flag–GFP–FIE protein was immunoprecipitated from 100 g transgenic seedlings at 8 days as described above (for Fig. 2e and Extended Data Fig. 4b). The Flag-GFP-FIE protein was separated and excised from a SDS–PAGE gel and subjected to in-gel digestion with trypsin. The phosphopeptides were enriched and analysed using the same LC–MS/MS method, except that targeted precursor ions and charge states (+2 and +3) were put in the inclusion list for targeted MS/MS acquisition. The accurate precursor mass and corresponding MS/MS spectra were validated using the synthetic phosphopeptide.
为了鉴定 FIE 中的 TOR 磷酸化位点,将 5 μg His-FIE 重组蛋白在激酶缓冲液中于 30 °C 下进行体外 TOR 激酶测定 2 小时。 磷酸化的 His-FIE 被 10% SDS-PAGE 凝胶分离。凝胶用 Thermo GelCode Blue Safe 蛋白质染料染色,并用 ddH2O 脱色。切除 His-FIE 条带并用胰蛋白酶进行凝胶内消化。使用来自 Glygen65 的 TiO2/ZrO2 填料富集磷酸肽,并使用 Orbitrap Fusion Tribrid 质谱仪 (Thermo Scientific)66 进行 LC-MS/MS 分析。LC-MS/MS谱图的原始数据由Mascot搜索引擎(2.2版本,Matrix Science)进行分析。参数为:拟南芥 TAIR10 数据库 (32,785 个条目,https://www.arabidopsis.org/)、10 ppm 的前体质量容差、0.8 Da 的片段质量容差、1 个缺失切割的胰蛋白酶、氧化 (M) 和磷酸化 (S、T、Y) 作为动态修饰。通过手动检查母离子质量和碎裂离子来确认鉴定的磷酸化肽和每个磷酸化位点分配。为了使用 Flag 抗体验证体内免疫沉淀的 FIE 磷酸化位点,如上所述,在 8 天从 100 g 转基因幼苗中免疫沉淀 Flag-GFP-FIE 蛋白(对于 Fig.2e 和扩展数据图4b). 从 SDS-PAGE 凝胶中分离并切除 Flag-GFP-FIE 蛋白,并用胰蛋白酶进行凝胶内消化。使用相同的 LC-MS/MS 方法富集和分析磷酸肽,但目标母离子和电荷态(+2 和 +3)被放入靶向 MS/MS 采集的包含列表中。 使用合成磷酸肽验证准确的母离子质量数和相应的 MS/MS 谱图。
Protoplast assays for microscopy detection, PUP-IT and amiRNA screening
用于显微镜检测、PUP-IT 和 amiRNA 筛选的原生质体检测
Mesophyll protoplasts were isolated from four-week-old WT leaves and transfected essentially as described67. For the PUP-IT proximity-tagging system (for Fig. 2a,b), protoplasts (2 × 105) in 1 ml MMg (0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7) were co-transfected with 100 μg of pUBQ10-TOR-C-PafA-HA-p2X35S-3×Flag-pup (E) and 100 μg of HBT–PRC2–MYC expressing different PRC2 components. Transfected protoplasts were incubated in 5 ml WI (0.5 M mannitol, 20 mM KCl, 4 mM MES, pH 5.7) in a 10 cm plate at room temperature for 12 h. The protoplast pellet was lysed in 1 ml extraction buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 0.5 mM DTT, and fresh protease inhibitor cocktail) with 0.1% SDS and subjected to immunoprecipitation using anti-Flag M2 Agarose Beads (Sigma, A2220) as described above. The input and immunoprecipitated proteins were mixed with 2× sample buffer and boiled for 10 min at 95 °C before gel electrophoresis and immunoblot analysis. For the confocal imaging of FIE and mutant proteins in mesophyll protoplasts (for Extended Data Fig. 5f), 200 μl protoplasts (4 × 104) were co-transfected with 38 μg of plasmid DNA to expression GFP-FIE or various mutants and 2 μg of pHBT-HY5–mCherry to express HY5–mCherry as a control for protoplast co-transfection and nuclear localization. Transfected cells were incubated in 1 ml WI in a 6-well plate at room temperature for 10 h before imaging with a Leica TCS SP8 laser scanning confocal microscopy (Leica). For optimal amiRNA screening (for Extended Data Fig. 7a), 200 μl protoplasts (4 × 104) were co-transfected with 35 μg of plasmid DNA expressing each candidate amiRNA (the empty amiRNA expression plasmid served as a control), 4 μg of pHSP-FIE-Flag plasmid, and 1 μg pHBT-GFP-HA plasmid (as a transfection internal control). Transfected cells were incubated in WI in a 6-well plate at room temperature for 3 h before the heat shock pulse at 37 °C for 1 h. Protoplasts were collected for immunoblot analysis after three more hours of incubation at room temperature.
从 4 周龄的 WT 叶中分离出叶肉原生质体,并基本上如所述转染67。对于 PUP-IT 接近标记系统(对于2a,b)、1 ml MMg(0.4 M 甘露醇、15 mM MgCl2、4 mM MES,pH 5.7)中的原生质体 (2 ×10 5) 与 100 μg 表达不同 PRC2 成分的 pUBQ10-TOR-C-PafA-HA-p2X35S-3×Flag-pup (E) 和 100 μg 表达不同 PRC2 成分的 HBT-PRC2-MYC 共转染。将转染的原生质体在 5 ml WI (0.5 M 甘露醇、20 mM KCl、4 mM MES,pH 5.7) 中在 10 cm 板中室温孵育 12 小时。将原生质体沉淀在 1 ml 提取缓冲液(25 mM Tris-HCl pH 7.6、150 mM NaCl、5 mM MgCl2、10% 甘油、0.1% NP-40、0.5 mM DTT 和新鲜蛋白酶抑制剂混合物)和 0.1% SDS 中裂解,并使用抗 Flag M2 琼脂糖珠(Sigma,A2220)进行免疫沉淀如上所述。将输入和免疫沉淀的蛋白质与 2× 样品缓冲液混合,并在 95 °C 下煮沸 10 分钟,然后进行凝胶电泳和免疫印迹分析。对于叶肉原生质体中 FIE 和突变蛋白的共聚焦成像(扩展数据图 .5f),将 200 μl 原生质体 (4 × 104) 与 38 μg 质粒 DNA 共转染以表达 GFP-FIE 或各种突变体,并用 2 μg pHBT-HY5–mCherry 共转染以表达 HY5–mCherry 作为原生质体共转染和核定位的对照。将转染的细胞在 6 孔板中的 1 ml WI 中室温孵育 10 小时,然后用 Leica TCS SP8 激光扫描共聚焦显微镜 (Leica) 成像。为了获得最佳 amiRNA 筛选(对于 Extended Data Fig. 7a),将 200 μl 原生质体 (4 × 104) 与 35 μg 表达每个候选 amiRNA 的质粒 DNA 共转染(空 amiRNA 表达质粒作为对照)、4 μg pHSP-FIE-Flag 质粒和 1 μg pHBT-GFP-HA 质粒(作为转染内参)。将转染的细胞在 WI 的 6 孔板中室温孵育 3 小时,然后在 37 °C 下热休克脉冲 1 小时。在室温下再孵育 3 小时后,收集原生质体用于免疫印迹分析。
Vernalization treatment Vernalization 治疗
For all the vernalization experiments (for Fig 4, Extended Data Fig. 9), plants were in or crossed into the FRISF2(Col-FRI or Ler-FRI) background42. Plants were grown on solid 0.5× MS (pH 5.7) medium (30 ml medium, 35 seedlings in 100 mm plates) or in liquid 0.5× MS (with 0.5% sucrose, pH 5.7) medium (1 ml medium, 6 seedlings in each well in 6-well-plates, or 10 ml medium, 50 seedlings in 100 mm plates) for chemical treatments under a 12 h light/12 h dark photoperiod condition at 23 °C/20 °C. For chemical treatments and GFP imaging (Fig. 4b–e and Extended Data Fig. 9a–d), liquid medium was used for more effective chemical penetration and handling intact roots for confocal imaging. For the other experiments, solid medium was used (Fig. 4a,h–j and Extended Data Fig. 9e). For glucose measurement, the results were more consistent using the solid medium for fresh weight measurement and tissue grinding. It was much easier to select the SSTS/AAAA/fie mutant when grown on a solid medium (Fig. 4i,j and Extended Data Fig. 9e). Notably, the results for gene expression and H3K27me3 dynamic during vernalization were similar when using liquid (Fig. 4b,e Extended Data Fig. 9b) or solid medium (Fig. 4i,j and Extended Data Fig. 9e). Plant samples collected at 8 days before vernalization were referred as NV. Seedlings were vernalized from the 8th day under an 8h light/16h dark short-day condition at 4 °C with 60% relative humidity and 40 μ mol m−2 s−1 light for 40 days (V), and then transferred back to the normal temperature condition (T) for seven more days for sample collection. For the tor-es mutant with the Col-FRI background, 10 μM oestradiol was added into the medium three days before vernalization. For Torin2 and INK128 treatment, 1 μM inhibitor was added in the medium one day before vernalization. Plant samples were collected every ten days during cold treatment, and medium with oestradiol or inhibitors were also refreshed every ten days. Collected samples were subjected to RNA expression quantification by RT–qPCR analysis and H3K27me3 quantification by ChIP–qPCR analysis. For glucose measurement during vernalization, plant materials were washed twice with cold water and frozen after removing all the liquid with Kimwipes. The glucose level was measured using the Glucose Assay Kit (Colorimetric) according to the manufacturer’s guidelines. For GUS histochemical staining, pFLC:FLC-GUS (Ler-FRI) seedlings before and after 40 days of vernalization were collected and stained essentially as described68. Flowering time was measured by counting rosette leaves at the time of bolting.
对于所有的春化实验(图 4,扩展数据图 3)。9),植物处于或杂交到 FRISF2 (Col-FRI 或 Ler-FRI) 背景中 42。植物生长在固体0.5× MS(pH 5.7)培养基(30 ml培养基,100 mm板中的35株幼苗)或液体0.×5 MS (含0.5%蔗糖,pH 5.7)培养基(1 ml培养基,6孔板中每个孔中6株幼苗,或10 ml培养基,100 mm板中的50株幼苗)中,在23°C/20°C下进行化学处理。 对于化学处理和 GFP 成像(图 D)。4b-e 和扩展数据图9a-d),液体介质用于更有效的化学渗透和处理完整的根以进行共聚焦成像。对于其他实验,使用固体培养基(图 D)。4a,h–j 和扩展数据图对于葡萄糖测量,使用固体培养基进行鲜重测量和组织研磨的结果更加一致。在固体培养基上生长时,选择 SSTS/AAAA/fie 突变体要容易得多(图 D)。4i,j 和扩展数据图值得注意的是,当使用液体时,春化过程中基因表达和 H3K27me3 动态的结果相似(图 D)。4B,E扩展数据 图9b) 或固体介质 (图 .4i,j 和扩展数据图9e). 在春化前 8 天收集的植物样品称为 NV。 从第 8 天开始,在 4 °C、60% 相对湿度和 40 μ mol m-2 s-1 光照条件下,将幼苗春化 40 天 (V),然后转移回常温条件 (T) 再 7 天进行样品采集。对于具有 Col-FRI 背景的 tor-es 突变体,在春化前 3 天向培养基中加入 10 μM 雌二醇。对于 Torin2 和 INK128 处理,在春化前 1 天在培养基中加入 1 μM 抑制剂。冷处理期间每 10 天收集一次植物样品,含雌二醇或抑制剂的培养基也每 10 天更新一次。通过 RT-qPCR 分析对收集的样品进行 RNA 表达定量,通过 ChIP-qPCR 分析对 H3K27me3 进行定量。对于春化过程中的葡萄糖测量,将植物材料用冷水洗涤两次,并在用 Kimwipes 去除所有液体后冷冻。根据制造商的指南,使用葡萄糖测定试剂盒(比色法)测量葡萄糖水平。对于 GUS 组织化学染色,收集春化 40 天前后的 pFLC:FLC-GUS (Ler-FRI) 幼苗,并基本如所述染色68。通过在抽薹时计算莲座叶来测量开花时间。
Protein blot analysis 蛋白质印迹分析
Plants were collected, frozen and ground into fine powder with a blue pestle in liquid nitrogen in a microfuge tube. For total protein extract, an equal volume of 2× SDS–PAGE protein loading buffer was added to each sample and boiled for 10 min. To detect histone proteins, nuclear extract enriched with histone was prepared as described69 and denatured by boiling in protein loading buffer. After centrifugation, the supernatant of each protein sample was separated by SDS–PAGE and transferred to the PVDF membrane for immunoblot analysis. Protein blots were probed using antibodies against TOR (1:1,000)21, pFIE(S14) (1:500), pS6K1(T449) (1:500)21, RPS6 (ref. 70) (1:5,000), pRPS6(S237) (1:3,000), pRPS6(S240)71 (1:3,000), H3 (Abcam, ab1791, 1:5,000), H3K4me3 (Abcam, ab8580, 1:1,000), H3K9me2 (Abcam 1220, 1:1,000), H3K27me3 (Millipore, 07–449, 1:1,000), H3K36me3 (Abcam, ab9050, 1:1,000), tubulin (Sigma, T6199, 1:5,000), GFP (CLONTECH, 632381, 1:5,000), HA–HRP (Sigma,12013819001,1:5,000), Flag–HRP (Sigma, A8592, 1:5,000), and MYC–HRP (Roche,1-814-150, 1:1,000), Anti-Mouse IgG–HRP (Sigma, A4416, 1:5000), Anti-Rabbit IgG–HRP (Sigma, A0545, 1:5,000). The signal intensity of each blot was quantified by ImageJ.
收集植物,冷冻并在微量离心管中用蓝色杵在液氮中研磨成细粉。对于总蛋白提取物,向每个样品中加入等体积的 2× SDS-PAGE 蛋白上样缓冲液并煮沸 10 分钟。为了检测组蛋白,如所述69 制备富含组蛋白的核提取物,并通过在蛋白质加载缓冲液中煮沸变性。离心后,通过 SDS-PAGE 分离每个蛋白质样品的上清液,并转移到 PVDF 膜上进行免疫印迹分析。使用抗 TOR (1:1,000)21、pFIE(S14) (1:) 的抗体探测蛋白质印迹 500)、pS6K1(T449) (1:500)21、RPS6(参考文献 70)(1:5,000)、pRPS6(S237)(1:3,000)、pRPS6(S240)71 (1:3,000)、H3(Abcam,ab1791,1:5,000)、H3K4me3(Abcam,ab8580,1:1,000)、H3K9me2(Abcam 1220,1:1,000)、H3K27me3(Millipore,07–449,1:1,000)、H3K36me3(Abcam,ab9050,1:1,000)、微管蛋白(Sigma,T6199,1:5,000)、GFP(CLONTECH,632381、 1:5,000)、HA-HRP(Sigma,12013819001,1:5,000)、Flag-HRP(Sigma,A8592,1:5,000)和 MYC-HRP(Roche,1-814-150,1:1,000),抗小鼠 IgG-HRP(Sigma,A4416,1:5000),抗兔 IgG-HRP(Sigma,A0545,1:5,000)。每个印迹的信号强度通过 ImageJ 量化。
Confocal microscopy 共聚焦显微镜
Transgenic plants expressing various GFP fusion proteins were grown in liquid 0.5× MS medium with 0.5% sucrose, pH 5.7 (for Fig 2i,j and Extended Data Fig. 6c,d). Transgenic plants expressing GFP–FIE or GFP–FIE(SSTS/AAAA) were grown without treatment for 4 days for image acquisition of leaf primordia and root elongation and root meristem zones. For TOR inhibitor treatment, plants were grown for 4 days and treated with 10 μM Torin2 or AZD8055 for 24 h before imaging. For plants in the tor-es mutant background, 10 μM oestradiol was added to the medium at the second day and incubated for 3 days before the image acquisition. For quantitative imaging of the dynamic GFP–FIE C/N ratio under sugar starvation, plants were geminated and grown in liquid 0.5× MS medium without sugar. Pictures were taken every day from day 2 to 6 (Extended Data Fig. 6f,g). For sugar starvation and glucose stimulation experiments (Extended Data Fig. 6h–j and Supplementary Video), plants were grown in liquid 0.5× MS medium without sugar for 5 days before glucose (25 mM) stimulation for 6 h before imaging. Plants for imaging were transferred into 35 mm glass bottom dish with 20 mm micro-well filled with 300 μl liquid 0.5× MS medium with 25 mM glucose and immediately used for time-lapse live imaging with 10 min intervals for 6 h. For imaging GFP–FIE and GFP-tagged FIE variants in protoplasts, transfected cells as previously described67 were incubated at room temperature for 10 h before imaging.
表达各种 GFP 融合蛋白的转基因植物在含有 0.5% 蔗糖的 0.5× MS 培养基液体中生长,pH 值为 5.7(图 2i,j 和扩展数据图 2)。6c,d)。表达 GFP-FIE 或 GFP-FIE(SSTS/AAAA)的转基因植物在未处理的情况下生长 4 天,用于获取叶原基和根伸长以及根分生组织区的图像。对于 TOR 抑制剂处理,植物生长 4 天,并在成像前用 10 μM Torin2 或 AZD8055 处理 24 小时。对于处于 tor-es 突变体背景下的植物,在第二天向培养基中加入 10 μM 雌二醇,并在图像采集前孵育 3 天。为了对糖饥饿下的动态 GFP-FIE C/N 比率进行定量成像,将植物成片并在不含糖的 0.5× MS 液体培养基中生长。从第 2 天到第 6 天每天拍摄照片(扩展数据图 D)。6f,g)。对于糖饥饿和葡萄糖刺激实验(扩展数据图6h-j 和补充视频),植物在不含糖的液体 0.5× MS 培养基中生长 5 天,然后在成像前进行葡萄糖 (25 mM) 刺激 6 小时。将用于成像的植物转移到 35 mm 玻璃底培养皿中,其中 20 mm 微孔中装有 300 μl 液体 0.5× MS 培养基和 25 mM 葡萄糖,并立即用于延时实时成像,间隔 10 分钟,持续 6 小时。为了对原生质体中的 GFP-FIE 和 GFP 标记的 FIE 变体进行成像,将如前所述67 个转染细胞在室温下孵育 10 小时,然后成像。
Confocal images were acquired using the Leica Application Suite X software on a Leica TCS SP8 (Leica) confocal microscope with the 20× objective lens for roots and protoplasts or the 40× water lens for leaf primordia. The scanning resolution was set to 1,024 Å × 1,024 or 2,048 Å × 2,048 pixels. To obtain fluorescence images of roots and protoplasts, the excitation was set at 488 nm (GFP) or 587 nm (mCherry) and emission at 494–541 nm (GFP) or 595–625 nm (mCherry). To obtain fluorescence images of leaf primordia, GFP and autofluorescence were visualized by excitation at 488 nm using the argon laser and emission at 510–520 nm (GFP) or 660–680 nm (autofluorescence). The autofluorescence signal was removed by applying the Channel and Spectral Dye Separation tool (Leica). For time-lapse live imaging, images were captured every 10 min for 6 h and replayed as a video. The images were collected and processed using Adobe Photoshop software. Quantification of subcellular localization of GFP-FIE were performed using LAS-X (Leica Application Suite v. X3.1.1.15751, Wetzlar, Germany). The GFP fluorescence intensity from the whole-cell and nuclear areas was measured. The fluorescence of the nuclear area was considered as GFP-FIE in the nucleus (N), and the fluorescence in the cytosol (C) was determined by subtracting the nucleus from the entire cell. The relative GFP fluorescent intensity between the cytosol (C) and nucleus (N) was analysed and presented as the quantitative C/N ratio from more than 15 cells in three biological replicates.
在 Leica TCS SP8 (Leica) 共聚焦显微镜上使用 Leica Application Suite X 软件采集共聚焦图像,该显微镜使用 20× 物镜用于根和原生质体,使用 40× 水镜用于叶原基。扫描分辨率设置为 1,024 Å × 1,024 或 2,048 Å × 2,048 像素。为了获得根和原生质体的荧光图像,激发波长设置为 488 nm (GFP) 或 587 nm (mCherry),发射波长为 494-541 nm (GFP) 或 595-625 nm (mCherry)。为了获得叶原基的荧光图像,使用氩激光在 488 nm 处激发并在 510-520 nm (GFP) 或 660-680 nm(自发荧光)处发射,从而可视化 GFP 和自发荧光。通过使用通道和光谱染料分离工具 (Leica) 去除自发荧光信号。对于延时实时成像,每 10 分钟捕获一次图像,持续 6 小时,并作为视频重播。这些图像是使用 Adobe Photoshop 软件收集和处理的。使用 LAS-X (Leica Application Suite v. X3.1.1.15751, Wetzlar, Germany) 对 GFP-FIE 的亚细胞定位进行定量。测量来自全细胞和核区域的 GFP 荧光强度。核区的荧光被认为是细胞核 (N) 中的 GFP-FIE,胞质溶胶 (C) 中的荧光是通过从整个细胞中减去细胞核来确定的。分析胞质溶胶 (C) 和细胞核 (N) 之间的相对 GFP 荧光强度,并以 3 个生物学重复中超过 15 个细胞的定量 C/N 比表示。
Processing and analysis of the ChIP–seq data
ChIP-seq 数据的处理和分析
ChIP–seq data were processed and analysed as previously described72. In brief, the quality of each sequencing library was assessed by examining fastq files with FastQC (version 0.11.5). Then, sequencing adaptors were removed using Trimmomatic (version 0.36)73. The Sickle program was used to eliminate bases with low quality scores (<20) and short reads (length < 20). The remaining clean reads were aligned to the reference sequence. The ChIP–seq reads were mapped to Arabidopsis (TAIR10) and human (hg19) combined genomes, using BWA-MEM (version 0.7.15-r1142-dirty). Moreover, the view function of samtools (version 1.6) was used to remove reads with quality (Q) < 20 (Q = −10log10 (p), where p is an estimate of the probability that the alignment does not correspond to the read’s true point of origin). The rmdup function of samtools (version 1.6) was used to remove duplicated reads mapped to exactly the same position because they were considered to be artifacts caused by PCR during the library construction step. The number of reads mapped to human genome was used to calculate reference derived normalization factor as previously described23. MACS274 utility callpeak was used to identify read-enriched regions (peaks) and utility pileup was used to generate bedgraph files with default settings. The number of reads in the bedgraphs was then scaled by the reference derived normalization factor to generate RRPM. The bedGraphToBigWig (version 4) of UCSC was used to transfer bedgraph files to bigwig files, and then the bigwig files were loaded onto JBrowse75 (version 1.12.1) for data visualization. Target genes were defined as genes with a peak within or near the gene body (±2 kb). For profiling the modification surrounding peak summits, regions from 2kb upstream and 2 kb downstream of peak summits were divided into 40 tiles in total (20 upstream tiles and 20 downstream tiles), the RRPM value in each title of each peak was used to plot heat map and the average RRPM value in each tile was used to make a line plot.
如前所述处理和分析 ChIP-seq 数据72。简而言之,通过使用 FastQC (版本 0.11.5) 检查 fastq 文件来评估每个测序文库的质量。然后,使用 Trimmomatic (版本 0.36)73 去除测序接头。使用 Sickle 程序去除质量评分低 (<20) 和短读长 (长度< 20) 的碱基。剩余的干净读长与参考序列对齐。使用 BWA-MEM (版本 0.7.15-r1142-dirty) 将 ChIP-seq 读长定位到拟南芥 (TAIR10) 和人类 (hg19) 联合基因组。此外,samtools(1.6 版)的视图函数用于删除质量 (Q) < 20 的读数(Q = −10log10 (p),其中 p 是比对与读数的真实原点不对应的概率的估计值)。samtools(1.6 版)的 rmdup 函数用于删除映射到完全相同位置的重复读数,因为它们被认为是文库构建步骤中由 PCR 引起的伪影。映射到人类基因组的读数数用于计算参考衍生的标准化因子,如前所述23。MACS274 实用程序 callpeak 用于识别读取丰富的区域 (峰),实用程序堆积用于生成具有默认设置的床头图文件。然后通过参考衍生的归一化因子对床图中的读数数进行缩放,以生成 RRPM。使用 UCSC 的 bedGraphToBigWig(版本 4)将床图文件传输到 bigwig 文件,然后将 bigwig 文件加载到 JBrowse75(版本 1.12.1)上进行数据可视化。 靶基因定义为在基因体内或附近具有峰 (±2 kb) 的基因。为了分析峰峰周围的变化,将峰峰上游 2kb 和下游 2 kb 的区域总共划分为 40 个瓦片(20 个上游瓦片和 20 个下游瓦片),每个峰每个标题中的 RRPM 值用于绘制热图,每个瓦片中的平均 RRPM 值用于制作线图。
Processing and analysis of the RNA-seq data
RNA-seq 数据的处理和分析
The RNA-seq reads were mapped to the Arabidopsis (TAIR10) genome with HISAT276 program (version 2.1.0). The featureCount program of Subread77 package (version 1.5.3) was used to quantify the RNA-seq expression levels of the TAIR10 annotated gene models. DESeq2 (ref. 78) was used to determine the significance of the differential expression between samples with the combined criteria: |log2 fold change| ≥ 1 and FDR ≤ 0.05. We added a pseudocount of 0.01 to DESeq2 normalized expression values to recalculate log2 fold change, which helped us avoid infinite values resulting from zero counts and seek some differentially expressed genes with relative low counts.
使用 HISAT276 程序 (2.1.0 版) 将 RNA-seq 读数定位到拟南芥 (TAIR10) 基因组。使用 Subread77 包 (版本 1.5.3) 的 featureCount 程序定量 TAIR10 注释基因模型的 RNA-seq 表达水平。DESeq2(参考文献 78)用于确定具有组合标准的样品之间差异表达的显着性: |log2 fold change|≥ 1 和 FDR ≤ 0.05。我们在 DESeq2 归一化表达值中添加了 0.01 的伪计数,以重新计算 log2 倍变化,这有助于我们避免由零计数产生的无限值,并寻找一些计数相对较低的差异表达基因。
Gene ontology term enrichment analysis
基因本体术语富集分析
The gene ontology (GO) term enrichment analysis was performed using The TAIR GO term enrichment tool BiNGO (http://www.psb.ugent.be/cbd/papers/BiNGO/Home.html)79. In brief, we first compiled the genes that are targeted by H3K27me3 and upregulated in STSS/AAAA/fie (Supplementary Table 5). Next, Fisher’s exact test (FDR < 0.05) was used by the website to identify GO terms that are significantly over-represented with the compiled gene list. Genes from ‘not assigned’ group were manually checked to identify the ones related to ‘regulation of transcription, DNA-templated (GO:0006355)’. The categories in biological process with fold enrichment > 2 and FDR < 10−10 were selected and presented in Extended Data Fig. 8b and Supplementary Table 6.
使用 TAIR GO 术语富集工具 BiNGO (http://www.psb.ugent.be/cbd/papers/BiNGO/Home.html)79 进行基因本体 (GO) 术语富集分析。简而言之,我们首先汇编了 H3K27me3 靶向并在 STSS/AAAA/fie 中上调的基因(补充表 5)。接下来,该网站使用 Fisher 精确检验 (FDR < 0.05) 来识别在编译的基因列表中明显过度代表的 GO 术语。手动检查来自“未分配”组的基因,以确定与“转录调节,DNA 模板化 (GO:0006355)”相关的基因。选择具有 2 倍富集和 FDR >< 10-10 的生物过程中的类别,并将其呈现在扩展数据图 中。8b 和补充表 6.
Chemical inhibitors for TOR kinase and S6K
TOR 激酶和 S6K 的化学抑制剂
The ATP-competitive chemical inhibitors for both mTORC1 and mTORC2, Torin2, AZD8055, INK128, WYE132 and WYE345, were tested empirically for efficacy in intact Arabidopsis seedlings and used in this study2,15,80,81,82,83,84,85. The second-generation ATP-competitive TOR kinase inhibitors, Torin2 and INK128, have been examined by kinome-wide selectivity profiles to ensure the specificity for TOR kinase inhibition80,81,82, and INK128 was shown to be effective in mammalian culture cells and in mice82. Torin2 was used for the initial chemical screens in seedlings up to 8-d treatment at different concentrations because Torin2 shows slower dissociation from TOR kinase and is more effective in sustained blocking of mTOR1 and mTORC2-mediated pAKT(S473) than AZD8055 in human cells84 and in Arabidopsis seedlings15. Chemical inhibitors for S6K, staurosporin and PF-4708671, were previously described4,86.
mTORC1 和 mTORC2、Torin2、AZD8055、INK128、WYE132 和 WYE345 的 ATP 竞争性化学抑制剂在完整拟南芥幼苗中的疗效进行了经验测试,并在本研究中使用 2,15,80,81,82,83,84,85。第二代 ATP 竞争性 TOR 激酶抑制剂 Torin2 和 INK128 已通过激酶组范围的选择性谱进行检查,以确保对 TOR 激酶抑制80,81,82 的特异性,并且 INK128 在哺乳动物培养细胞和小鼠中被证明有效82。Torin2 用于不同浓度下长达 8 天的幼苗的初始化学筛选,因为 Torin2 与 TOR 激酶的解离速度较慢,并且在持续阻断 mTOR1 和 mTORC2 介导的 pAKT (S473) 方面比AZD8055在人细胞84 和拟南芥幼苗中更有效15。S6K 的化学抑制剂、星形孢菌素和 PF-4708671 之前已描述过 4,86。
Statistics 统计学
For most of the quantitative data shown in this paper represent mean ± s.d. from at least three biological replicates. Statistical analyses were performed with GraphPad Prism 9 software. Unless otherwise specified, statistical significance was determined with the unpaired two-tailed Student’s t-test when comparing two groups and one-way or two-way ANOVA when comparing more than two groups with Tukey’s multiple comparisons test. The exact P values are indicated in the figures.
对于本文所示的大多数定量数据,代表至少三次生物学重复的平均 ± s.d.。使用 GraphPad Prism 9 软件进行统计分析。除非另有说明,否则在比较两组时用未配对的双尾 Student's t 检验确定统计显着性,当用 Tukey 的多重比较检验比较两组以上时,用单向或双向方差分析确定统计显着性。确切的 P 值如图所示。
Reporting summary 报告摘要
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
有关研究设计的更多信息,请参阅本文链接的 Nature Research 报告摘要。
Data availability 数据可用性
Sequencing data have been deposited to the Gene Expression Omnibus under accession GSE161807. The pCambia-PUP-IT vector was deposited to Addgene (#186478) .The plasmids and the transgenic Arabidopsis seeds generated in this study are available upon request. Source data are provided with this paper.
测序数据已根据 Accession GSE161807 存入 Gene Expression Omnibus。将 pCambia-PUP-IT 载体沉积到 Addgene (#186478) 中。本研究中生成的质粒和转基因拟南芥种子可应要求提供。源数据随本文提供。
Code availability 代码可用性
Analysis codes are available upon reasonable request from the corresponding authors. No custom codes were central to the conclusions of the paper.
分析代码可应通讯作者的合理要求提供。没有自定义代码是论文结论的核心。
References 引用
Krejci, A. & Tennessen, J. M. Metabolism in time and space–exploring the frontier of developmental biology. Development 144, 3193–3198 (2017).
Krejci, A. & Tennessen, J. M. 时间和空间的新陈代谢——探索发育生物学的前沿。发展144, 3193–3198 (2017)。Shi, L., Wu, Y. & Sheen, J. TOR signaling in plants: conservation and innovation. Development 145, dev160887 (2018).
Shi, L., Wu, Y. & Sheen, J. 植物中的 TOR 信号:保护和创新。发展145, dev160887 (2018)。Li, L., Liu, K. H. & Sheen, J. Dynamic nutrient signaling networks in plants. Annu. Rev. Cell Dev. Biol. 37, 341–367 (2021).
Li, L., Liu, K. H. & Sheen, J. 植物中的动态营养信号网络。Annu. Rev. Cell Dev. Biol.37, 341–367 (2021 年)。Xiong, Y. et al. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013).
Xiong, Y. 等人。葡萄糖-TOR 信号转导重编程转录组并激活分生组织。自然496, 181–186 (2013)。Dobrenel, T. et al. TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 67, 261–285 (2016).
Dobrenel, T. et al. TOR 信号传导和营养传感。植物生物学年鉴。67, 261–285 (2016 年)。Mozgova, I. & Hennig, L. The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296 (2015).
Mozgova, I. & Hennig, L.多梳组蛋白质调节网络。植物生物学年鉴。66, 269–296 (2015 年)。Pu, L. & Sung, Z. R. PcG and trxG in plants—friends or foes. Trends Genet. 31, 252–262 (2015).
Pu, L. & Sung, Z. R. 植物中的PcG和trxG——朋友或敌人。趋势基因。31, 252–262 (2015 年)。Bieluszewski, T., Xiao, J., Yang, Y. & Wagner, D. PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci. 26, 1186–1198 (2021).
Bieluszewski, T., Xiao, J., Yang, Y. & Wagner, D. PRC2 活性、募集和沉默:比较视角。趋势植物科学。26, 1186–1198 (2021)。Atlasi, Y. & Stunnenberg, H. G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18, 643–658 (2017).
Atlasi, Y. & Stunnenberg, H. G.干细胞分化和发育过程中表观遗传标记的相互作用。Nat. Rev. Genet.18, 643–658 (2017 年)。Schuettengruber, B., Bourbon, H. M., Di Croce, L. & Cavalli, G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57 (2017).
Schuettengruber, B., Bourbon, H. M., Di Croce, L. & Cavalli, G. 通过多梳和三胸进行基因组调控:70 年并且还在增加。单元格171, 34–57 (2017)。Margueron, R. et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Margueron, R. 等人。多梳蛋白 EED 在抑制组蛋白标记传播中的作用。自然461, 762–767 (2009)。Bouyer, D. et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7, e1002014 (2011).
Bouyer, D. 等人。Polycomb repressive complex 2 控制胚胎到幼苗的相变。PLoS 基因。7, e1002014 (2011).Bernier, G. & Perilleux, C. A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3, 3–16 (2005).
Bernier, G. & Perilleux, C.开花时间控制遗传学的生理学概述。植物生物技术杂志3, 3–16 (2005 年)。Yu, S., Lian, H. & Wang, J. W. Plant developmental transitions: the role of microRNAs and sugars. Curr. Opin. Plant Biol. 27, 1–7 (2015).
Yu, S., Lian, H. & Wang, JW 植物发育转变:microRNA和糖的作用。电流。意见。植物生物学。27, 1-7 (2015 年)。Li, X. et al. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl Acad. Sci. USA 114, 2765–2770 (2017).
Li, X. 等人。拟南芥根和芽顶端的差异 TOR 激活和细胞增殖。美国国家科学院院刊 114, 2765–2770 (2017)。Barbier, F. F., Dun, E. A., Kerr, S. C., Chabikwa, T. G. & Beveridge, C. A. An update on the signals controlling shoot branching. Trends Plant Sci. 24, 220–236 (2019).
Barbier, F. F., Dun, E. A., Kerr, S. C., Chabikwa, T. G. & Beveridge, C. A.控制拍摄分支的信号的更新。趋势植物科学。24, 220–236 (2019 年)。Deprost, D. et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8, 864–870 (2007).
Deprost, D. 等人。拟南芥 TOR 激酶将植物生长、产量、抗逆性和 mRNA 翻译联系起来。EMBO Rep.8, 864–870 (2007)。Ryabova, L. A., Robaglia, C. & Meyer, C. Target of Rapamycin kinase: central regulatory hub for plant growth and metabolism. J. Exp. Bot. 70, 2211–2216 (2019).
Ryabova, L. A., Robaglia, C. & Meyer, C. 雷帕霉素激酶的目标:植物生长和代谢的中心调节枢纽。J. Exp. Bot.70, 2211–2216 (2019)。Wu, Y. et al. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 70, 2227–2238 (2019).
Wu, Y. 等人。通过 TOR 在植物中整合营养、能量、光和激素信号。J. Exp. Bot.70, 2227–2238 (2019 年)。Ren, M. et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24, 4850–4874 (2012).
任, M. et al.雷帕霉素信号转导的靶标调节拟南芥的代谢、生长和寿命。植物细胞24, 4850–4874 (2012)。Xiong, Y. & Sheen, J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287, 2836–2842 (2012).
Xiong, Y. & Sheen, J. 植物中雷帕霉素和雷帕霉素 (TOR) 蛋白信号传导的葡萄糖靶标。J. Biol. 化学。287, 2836–2842 (2012 年)。Caldana, C. et al. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 73, 897–909 (2013).
Caldana, C. 等人。雷帕霉素突变体诱导靶标的全身分析揭示了控制拟南芥生长的一般代谢开关。J 工厂73, 897–909 (2013 年)。Orlando, D. A. et al. Quantitative ChIP–seq normalization reveals global modulation of the epigenome. Cell Rep. 9, 1163–1170 (2014).
Orlando, DA et al. 定量 ChIP-seq 标准化揭示了表观基因组的整体调节。细胞代表 9, 1163–1170 (2014)。Anderson, G. H., Veit, B. & Hanson, M. R. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3, 12 (2005).
Anderson, G. H., Veit, B. & Hanson, M. R.Arabidopsis AtRaptor 基因对于胚胎后植物生长至关重要。BMC 生物学。3, 12 (2005 年)。Moreau, M. et al. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481 (2012).
Moreau, M. 等人。LST8/GbetaL 的拟南芥同源物是雷帕霉素激酶靶标的伴侣,其突变会损害植物的生长、开花和对长日照的代谢适应。植物细胞24, 463–481 (2012)。Forzani, C. et al. Mutations of the AtYAK1 kinase suppress TOR deficiency in Arabidopsis. Cell Rep. 27, 3696–3708.e3695 (2019).
Forzani, C. 等人。AtYAK1 激酶的突变抑制拟南芥中的 TOR 缺陷。细胞代表 27, 3696–3708.e3695 (2019)。Liu, Q. et al. A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 15, 715–722 (2018).
Liu, Q. 等人。用于识别膜蛋白-蛋白质相互作用的邻近标记系统。Nat. Methods15, 715–722 (2018)。Kallberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012).
Kallberg, M. 等人。使用 RaptorX Web 服务器进行基于模板的蛋白质结构建模。Nat. Protoc.7, 1511–1522 (2012 年)。Tie, F., Siebold, A. P. & Harte, P. J. The N-terminus of Drosophila ESC mediates its phosphorylation and dimerization. Biochem. Biophys. Res. Commun. 332, 622–632 (2005).
Tie, F., Siebold, A. P. & Harte, P. J.果蝇 ESC 的 N 端介导其磷酸化和二聚化。生化。生物物理学。Res. Commun.332, 622–632 (2005 年)。Oliva, M. et al. FIE, a nuclear PRC2 protein, forms cytoplasmic complexes in Arabidopsis thaliana. J. Exp. Bot. 67, 6111–6123 (2016).
Oliva, M. et al. FIE 是一种核 PRC2 蛋白,在拟南芥中形成细胞质复合物。J. Exp. Bot.67, 6111–6123 (2016 年)。Katz, A., Oliva, M., Mosquna, A., Hakim, O. & Ohad, N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37, 707–719 (2004).
Katz, A., Oliva, M., Mosquna, A., Hakim, O. & Ohad, N. FIE和CURLY LEAF多梳蛋白在孢子体发育过程中相互作用调节同源框基因表达。J 工厂37, 707–719 (2004 年)。Zhang, X. et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5, e129 (2007).
Aichinger, E., Kornet, N., Friedrich, T. & Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 63, 615–636 (2012).
Pierre-Jerome, E., Drapek, C. & Benfey, P. N. Regulation of division and differentiation of plant stem cells. Annu. Rev. Cell Dev. Biol. 34, 289–310 (2018).
Denyer, T. et al. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48, 840–852.e845 (2019).
Liu, C., Thong, Z. & Yu, H. Coming into bloom: the specification of floral meristems. Development 136, 3379–3391 (2009).
Wang, L. et al. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583, 277–281 (2020).
Petricka, J. J., Clay, N. K. & Nelson, T. M. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 56, 251–263 (2008).
Yang, Y. et al. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 14, e1007296 (2018).
Menon, G., Schulten, A., Dean, C. & Howard, M. Digital paradigm for Polycomb epigenetic switching and memory. Curr. Opin. Plant Biol. 61, 102012 (2021).
Ruelens, P. et al. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4, 2280 (2013).
Yang, H. et al. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357, 1142–1145 (2017).
Questa, J. I., Song, J., Geraldo, N., An, H. & Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353, 485–488 (2016).
Purvis, O. N. Vernalization of fragments of embryo tissue. Nature 145, 462 (1940).
Klotke, J. K., Gatzke, J. & Heyer, N. A.G. Impact of soluble sugar concentrations on the acquisition of freezing tolerance in accessions of Arabidopsis thaliana with contrasting cold adaptation—evidence for a role of raffinose in cold acclimation. Plant Cell Environ. 27, 1395–1404 (2004).
Zhao, Y., Antoniou-Kourounioti, R. L., Calder, G., Dean, C. & Howard, M. Temperature-dependent growth contributes to long-term cold sensing. Nature 583, 825–829 (2020).
Heo, J. B. & Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79 (2011).
Xiao, J. et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552 (2017).
Zhou, Y. et al. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 50, 638–644 (2018).
Dong, Y. et al. TOR represses stress responses through global regulation of H3K27 trimethylation in plants. Preprint at bioRxiv https://doi.org/10.1101/2021.03.28.437410 (2021).
Wang, H. et al. Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of Polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet. 12, e1005771 (2016).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Li, J. F. et al. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25, 1507–1522 (2013).
Brand, L. et al. A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol. 141, 1194–1204 (2006).
Liu, K. H. et al. Discovery of nitrate–CPK–NLP signalling in central nutrient-growth networks. Nature 545, 311–316 (2017).
Shu, J. et al. Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct 3, e00100 (2019).
Schubert, D. et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25, 4638–4649 (2006).
de Lucas, M. et al. Transcriptional regulation of Arabidopsis Polycomb Repressive Complex 2 coordinates cell-type proliferation and differentiation. Plant Cell 28, 2616–2631 (2016).
Angel, A., Song, J., Dean, C. & Howard, M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108 (2011).
Ori, N., Eshed, Y., Chuck, G., Bowman, J. L. & Hake, S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532 (2000).
Arur, S. & Schedl, T. Generation and purification of highly specific antibodies for detecting post-translationally modified proteins in vivo. Nat. Protoc. 9, 375–395 (2014).
Niu, Y. & Sheen, J. Transient expression assays for quantifying signaling output. Methods Mol. Biol. 876, 195–206 (2012).
Yamaguchi, N. et al. Chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12, e0170 (2014).
Jacob, Y. et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253 (2014).
Zhang, T., Schneider, J. D., Zhu, N. & Chen, S. Identification of MAPK substrates using quantitative phosphoproteomics. Methods Mol. Biol. 1578, 133–142 (2017).
Pang, Q. et al. Proteomics and phosphoproteomics revealed molecular networks of stomatal immune responses. Planta 252, 66 (2020).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Cervera, M. Histochemical and fluorometric assays for uidA (GUS) gene detection. Methods Mol. Biol. 286, 203–214 (2005).
Ye, R. et al. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell 46, 859–870 (2012).
Chen, G. H., Liu, M. J., Xiong, Y., Sheen, J. & Wu, S. H. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc. Natl Acad. Sci. USA 115, 12823–12828 (2018).
Enganti, R. et al. Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Front. Plant Sci. 8, 2210 (2017).
Li, Z. et al. The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol. 20, 139 (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS).Genome Biol. 9, R137 (2008).
Buels, R. et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 17, 66 (2016).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Liu, Q. et al. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J. Med. Chem. 54, 1473–1480 (2011).
Liu, Q. et al. Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae. ACS Chem. Biol. 7, 982–987 (2012).
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Liu, Q. et al. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J. Biol. Chem. 287, 9742–9752 (2012).
Liu, Q. et al. Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 73, 2574–2586 (2013).
Montane, M. H. & Menand, B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64, 4361–4374 (2013).
Pearce, L. R. et al. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1). Biochem. J. 431, 245–255 (2010).
Acknowledgements
We thank A. von Arnim, D. Anwesha, S. H. Wu, and C. Meyer for sharing pRPS6(S237), pRPS6(S240) and RPS6 antibodies with detailed protocols; C. Meyer, J. Brunkard, C. Dean, D. Bouyer, Y. H. Cui, J. Goodrich, S. Brady, M. de Lucas and ABRC for providing Arabidopsis lines; H. Y. Qi for supplying the human 293T cells; J. Bush for plant management; S. Jiang and F. Marchan for help with insect cell expression system; B. Ardehali, C. Tsokos, F. K. Hsieh and C. H. Yang for sharing reagents and discussions; C. Dufresne from Thermo Scientific Training Institute for advice on LC–MS/MS analysis; X. Fang, A. Diener, L. Li, J. Bush, T. C. Chen and H. Y. Cho for critical reading of the manuscript. This work was supported by the NIH grants GM060493 and GM129093 to J. Sheen and R.Y., the Agriculture and Food Research Initiative (2020-67013-31615) from the USDA-NIFA to S. Chen, and National Natural Science Foundation of China (31770285) to Y.Z.
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 TOR controls the global H3K27me3 level and development.
a, TOR differentially regulates seedling biomass. WT seedlings treated with different concentrations of Torin2 or tor-es seedlings at 8 days after germination. (n = 6 seedlings). b, Distinct TOR activity thresholds regulate DNA replication. Quantification of EdU staining in roots (n = 5 seedlings). c, Metaplots showing ChIP-Rx-seq read density. H3K27me3 and H3K9me2 in 7-day WT and tor-es. The ChIP-seq data are normalized with an exogenous reference genome. The peak summits ±2 kb is shown. d, Genome browser view of H3K27me3 and H3K9me2 ChIP-seq read densities in WT and tor-es. The Arabidopsis genome region within Chr3:15900000..17800000 is shown. An enlarged view of a selected region is shown on the top. e, The H3K27me3 level in plants grown in sugar-containing medium. d, day. Values are the relative level of H3K27me3 compared with the corresponding H3 control, with immunoblot signals in day3 set as 1.0. Experiments were conducted in three biological repeats with similar results. Data in a, b show mean ± s.d., one-way ANOVA with Tukey’s multiple comparisons test.
Extended Data Fig. 2 TOR controls the global H3K27me3 level.
a-d, TOR regulates global H3K27me3 levels. a, TOR but not S6K regulates global H3K27me3 levels. WT or S6K1-HA transgenic seedlings were treated with 10 μM of different inhibitors for 3 days. At 7 days, TOR activity was monitored by pS6K1(T449) and the band shift of pS6K1-HA. S6K activity was monitored by pRPS6(S237) and pRPS6(S240). The intensity of each immunoblot was quantified by ImageJ. Values are the relative level of H3K27me3 compared with the corresponding H3 control, with immunoblots in mock set as 1.0. b, Heatmap of H3K27me3 enrichment in plants with or without TOR inhibitor treatment. The colour scale indicates reference-adjusted RPM (RRPM) surrounding peak summit from the ChIP-Rx-seq data. c, Metaplots showing H3K27me3 ChIP-Rx-seq read density in plants with or without TOR inhibitor treatment. The ChIP-seq data are normalized with an exogenous reference genome. The peak summits ±2 kb is shown. d, Genome browser view of H3K27me3 ChIP-seq read densities. The Arabidopsis genome region within Chr3:15800000..18000000 is shown. e, Differential regulation of S6K and H3K27me3 in rap1 and lst8-1 mutants. The restoration of H3K27me3 was induced by 25 mM glucose for 6 h in 7-d sugar-starved seedlings. TOR-S6K activity was monitored by pRPS6(S237) and pRPS6(S240). The intensity of each immunoblot was quantified by ImageJ. Values for H3K27me3 are the relative level of H3K27me3 compared with the corresponding H3 control, with immunoblots in WT before glucose stimulation set as 1.0. Values for RPS6 phosphorylation are the relative level of pRPS6(S237) and pRPS6(S240) compared with the corresponding RPS6 control, with immunoblots in WT after glucose stimulation set as 1.0. Data in a and e are representatives of three biological replicates each.
Extended Data Fig. 3 The transcript and protein levels of PRC2 components are not regulated by TOR.
a, Evolutionarily conserved core PRC2 subunits in Arabidopsis, Drosophila and mammals. PRC2 components regulating plant postembryonic development are shown. b, RT-qPCR analysis of genes encoding PRC2 components in 7-day WT and tor-es. ACT2 transcripts served as an internal control for normalization. Data show mean ± s.d. from 4 biological replicates. Data were analysed by unpaired two-sided Student’s t test. c, GFP-tagged PRC2 components are not regulated by TOR. Torin2 (10 μM) was added for 24 h in 7-d seedlings. Tubulin was used for the loading control for the immunoblot analyses. Experiments were conducted in three biological repeats with similar results.
Extended Data Fig. 4 TOR directly interacts with and phosphorylates FIE.
a, Glucose enhances the interaction between TOR and FIE in vivo. Co-immunoprecipitation (Co-IP) of FLAG-tagged FIE with TOR from starved (7 d) and glucose stimulated (2 h) plants. Glc, Glucose. b, Summary of the TOR phosphorylation sites of FIE identified by LC-MS/MS analysis. The corresponding phosphopeptides and its covered regions are listed. Expected, the expect value indicates the probability that the peptide is matched by chance. Smaller value indicates more significance of the peptide identification. Score, the score value represents a calculation of how well the observed spectrum fits to the identified peptide. Higher value indicates higher confidence of the peptide identification. No. of matches, the total number of matched peptides with the same modifications and sites from three biological repeats. Validated in vivo, the modified peptides were identified from in vivo FLAG-FIE immunoprecipitation. c, Mass spectrometric analysis of pS10 peptide from in vitro TOR kinase assay. d, Mass spectrometric analysis of pS14 peptide from in vitro TOR kinase assay. e, Mass spectrometric analysis of pT16 peptide from in vitro TOR kinase assay. f, Mass spectrometric analysis of pS18 peptide from in vivo FLAG-FIE immunoprecipitation. g, The predicted FIE structure. The N-terminal 34 aa sequence is shown with phosphorylation sites (red) identified by mass spectrometry and the basic residues (blue). The predicted 3D structure of FIE by modelling is shown with the flexible N-terminal domain highlighted in green (right). h, TOR phosphorylation of FIE variants by in vitro kinase assays. Single or quadruple mutants of FIE protein was used as the substrate. Phosphorylation of His-FIE by TOR is shown with autoradiography (top). Protein loading control is shown by Coomassie blue staining (bottom). Experiments were conducted in three biological repeats with similar results.
Extended Data Fig. 5 Conservation of the key TOR phosphorylation sites in plant FIE and animal orthologs.
a, Phosphosite conservation was analysed by multiple sequence alignment of FIE proteins from reference organisms using PLAZA 4.0 Dicots (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_dicots/). The selection of reference organisms includes Arabidopsis, Brassica, soybean, rice, maize and wheat. The four arrow heads indicate the potential phosphorylation residues. b, N-terminal sequences of Drosophila ESC (residues 1–60) and human EED (residues 21–80) are shown. Highly conserved positions of S/T-rich and basic regions are indicated. c, Immunoblot validation of pFIE(S14) specific antibody using the in vitro TOR-FIE kinase assay. d, Glucose enhances pFIE(S14) levels. Immunoblot analysis of pFIE(S14) after IP with anti-FLAG in starved (7 d) transgenic FLAG-GFP-FIE seedlings and stimulated by different concentrations of glucose for 2 h. e, GFP-FIE protein levels are not regulated in tor-es or Torin2 treated 7-d seedlings. A specific TOR antibody was used to detect endogenous TOR by immunoblot analysis. Tubulin served as the loading control. Values are the relative level of GFP-FIE over Tubulin, with blots in mock treatment set as 1.0. f, In vitro histone methyltransferase assays using H3 substrate and recombinant Arabidopsis PRC2 complexes from insect Sf9 cells. The complexes stained by Coomassie blue were purified with FLAG-tagged WT or the mutant form (SSTS/AAAA) of FIE. The * indicates a nonspecific protein from insect cells. The H3K27me3 was detected by immunoblot and quantified by comparing to the corresponding H3 control.
Extended Data Fig. 6 Glucose-TOR specifically promotes the cytoplasm-to-nucleus translocation of FIE to enhance the PRC2 activity in the nucleus.
a, A proposed model for the cytoplasm-to-nucleus translocation of FIE regulated by TOR. FIE mainly localizes in the cytoplasm at low TOR activity and its phosphorylation by glucose-activated TOR stimulates its nuclear entry to enhance PRC2 activity. Blocking TOR activity or mutation of the phosphorylation sites inhibited the nuclear translocation of FIE. b, Quantitative confocal imaging of GFP-FIE and GFP-FIE(SSTS/AAAA) in leaf primordia and roots. The cytoplasm/nucleus (C/N) signal intensity ratio of GFP-FIE or GFP-FIE(SSTS/AAAA) at the single-cell level was measured by quantitative confocal imaging using the Leica LAS-X software. WT seedlings (5 d) expressing GFP-FIE without (Ctrl) or with 10 μM of Torin2 or AZD treatment (24 h) or tor-es (10 μM estradiol for 3 d) were examined. Root elongation zone and root meristem zone are illustrated. c, Confocal images of GFP-tagged PRC2 components in the meristem zone of roots. Plants were imaged with or without (Mock) Torin2 treatment. d, Confocal images of GFP-tagged FIE and mutants in protoplasts. BF, bright field. NLS–mC denotes nuclear HY5–mCherry as a control for protoplasts co-transfection and nuclear localization. Scale bars, 10 μm. Images are representative of 10 protoplasts from three biological repeats. e, Immunoblot analysis of GFP-tagged FIE variants expressed in protoplast and transgenic plants. Tubulin was used for the loading control. Data are representatives of three biological replicates. f, g, The dynamics of GFP-FIE during glucose starvation. f, Confocal images of GFP-FIE from 2–6 d in the root elongation zone in glucose-free medium. g, Quantitative confocal imaging of GFP-FIE C/N ratio. h, i, j, Glucose stimulates dynamic nuclear translocation of GFP-FIE after starvation in the root elongation zones. h, Confocal images of GFP-FIE without or with 25 mM glucose stimulation for 6 h in starved seedlings (5 d). i, Quantitative confocal imaging of GFP-FIE C/N ratio. j, Time-lapse live imaging of glucose stimulated nuclear translocation of GFP-FIE in the root elongation zone. Representative images were taken from Supplementary Video at 1–4 h time points after 25 mM glucose stimulation. The experiment was repeated three times with similar results. b, f, i, In the boxplots, data were analysed from more than 15 cells from three experiments, and are expressed as mean ± s.d. Centre line, median; box limits, 25th and 75th percentiles; the whiskers indicate data's minimum and maximum; the points represent each individual value. Statistical significance was determined by unpaired two-sided Student’s t test. c, g, h, j, Images are representative of six seedlings from three biological repeats. Scale bar, 25 μm.
Extended Data Fig. 7 Generation and characterization of estradiol-inducible fie-amiR-es and SSTS/AAAA/fie mutants.
a, Screening of optimal amiRNA in the protoplast system. Empty amiRNA expression vector as a control (Ctrl) or each of the eight amiRNA candidate plasmids was co-transfected with a heat shock promoter-driven FIE-FLAG plasmid. The HBT-GFP-HA plasmid serves as an internal control for all co-transfection experiments. Immunoblot analysis indicated similar GFP-HA protein levels. The most potent amiRNA4 abolished FIE-FLAG expression and was chosen to generate estradiol-inducible fie-amiR-es transgenic plants. b, RT-qPCR analysis of FIE transcripts in 14-day transgenic fie-amiR-es lines without or with 10 μM of estradiol treatment. UBQ10 transcripts served as an internal control for normalization. Data show mean ± s.d. from 3 biological replicates. Data were analysed by unpaired two-sided Student’s t test. c, Protein blot analysis of FIE protein levels in 14-day transgenic fie-amiR-es lines. Three independent fie-amiR-es lines were crossed to the FLAG-GFP-FIE transgenic plant. The FLAG-GFP-FIE protein was eliminated with 10 μM of estradiol treatment. Tubulin was used for the loading control. d, The development phenotype of the fie-amiR-es lines was similar to that of the SSTS/AAAA/fie mutant. Three independent fie-amiR-es lines and the SSTS/AAAA/fie mutant showed small, narrow and curled leaves. The GFP-FIE/fie plant showed normal development. Scale bar, 10 mm. Experiments were conducted in three biological repeats with similar results. e, The H3K27me3 level was greatly decreased in 14-day fie mutant plants. Values are the relative level of H3K27me3 compared with the corresponding H3 control, with immunoblot signals in WT set as 1.0. f, Genome browser view of H3K27me3 ChIP-seq read densities. The Arabidopsis genome region within Chr3:15800000..18000000 is shown. g, Venn diagram of H3K27me3 target genes in Arabidopsis from three independent genome wide analyses. The H3K27me3 targets in our study cover 79.7% of genes from Zhang et al. and 84.5% of genes from Bouyer et al12. Data in a, c, and e are representatives of three biological replicates each.
Extended Data Fig. 8 The TOR-FIE-PRC2-TR relay plays a central role in diverse developmental programs.
a, TOR-FIE-PRC2 target genes. These genes are marked by H3K27me3 and upregulated in the SSTS/AAAA/fie mutant. b, Gene Ontology enrichment analysis of TOR-FIE-PRC2 target genes in biological process terms related to transcriptional regulation and development. Fisher’s exact test was used by the BiNGO to identify GO terms that are significantly over-represented with the compiled gene list. FDR, false discovery rate. The categories in biological process with fold enrichment > 2 and FDR < 10−10 were selected and presented. c, PRC2 target genes are transcription regulators upregulated in fie mutants (14d). UBQ10 or ACT2 transcripts served as an internal control for normalization. d, f, RT-qPCR analysis of TOR-FIE-PRC2 target genes in WT and tor-es. (d) In sugar-containing medium or (f) in sugar-free medium and without or with 25 mM glucose stimulation for 6 h after starvation (7 d). UBC21 transcripts served as an internal control for normalization. e, g, ChIP-qPCR analysis of H3K27me3 enrichment on TOR-FIE-PRC2 target genes in 7-day WT and tor-es. (e) in sugar-containing medium or (g) in sugar-free medium and without and with 25 mM glucose stimulation for 6 h after starvation (7 d). ChIP-qPCR data were normalized to percentage of input DNA. c-g, Data show mean ± s.d. from 3 biological replicates. Data were analysed by unpaired two-sided Student’s t test.
Extended Data Fig. 9 VIN3 expression, H3K27me3 enrichment at FLC, and GFP-FIE nuclear translocation during vernalization.
a, VIN3 Induction by vernalization is not regulated by TOR. Seedlings were treated at 4 °C to induce vernalization for the indicated days without (Ctrl) or with TOR inhibitors (1 μM Torin2 and INK128), or in the inducible tor-es mutant. Data are relative to the UBC21 (At5g25760) expression as a control gene and normalized to non-vernalized (NV) levels of ctrl plants. Data show mean ± s.d. from 3 biological replicates, two-way ANOVA with Tukey’s multiple comparisons test. b, TOR deficiency reduces H3K27me3 levels at FLC. Relative H3K27me3/H3 level normalized to that of ctrl plants under non-vernalized (NV) condition as 1. Black boxes, exons. Red boxes, selected regions for H3K27me3 ChIP-qPCR analyses. bp, base pairs. FLC-3 and FLC-4,5 are located in the nucleation and spreading regions of H3K27me3 at FLC, respectively. c, d, Prolonged cold treatment prompts the nuclear localization of GFP-FIE in the elongation zones of roots. c, Confocal images of GFP-FIE during vernalization. Images are representative of six seedlings from three biological repeats. Scale bar, 25 μm. d, Quantitative confocal imaging of GFP-FIE in the root elongation zone. The cytoplasm/nucleus (C/N) signal intensity ratio of GFP-FIE at the single-cell level was measured by quantitative confocal imaging using the Leica LAS-X software. In the boxplot, data were analysed from more than 15 cells from three experiments, and are expressed as mean ± s.d. Centre line, median; box limits, 25th and 75th percentiles; the whiskers indicate data's minimum and maximum; the points represent each individual value. Statistical significance was determined by unpaired two-sided Student’s t test. e, Vernalization induced H3K27me3 levels at FLC are compromised in AAAA/fie-FRI. Relative H3K27me3/H3 level was normalized to that of fie/Col-FRI plants under non-vernalized (NV) condition as 1. Data in b, e show mean ± s.d. from 3 biological replicates, two-way ANOVA with Tukey’s multiple comparisons test; NV, non-vernalized. V, vernalization days at 4 °C. T, postcold days at 22 °C.
Extended Data Fig. 10 Model of the Glucose-TOR-FIE-PRC2 signalling network governing diverse developmental programs.
a, The Glucose-TOR-FIE-PRC2 signalling network. Optimal photosynthesis in source leaves produces sugars that are transported to energy demanding sinks, including apical and lateral meristems, developing leaf primordia and young leaves, roots, flowers, fruits and seeds, to support their growth and development. In the sink, glucose derived from local or systemic carbon sources is metabolized to activate TOR kinase, which interacts with and phosphorylates FIE in the cytoplasm. The phosphorylated FIE is translocated into the nucleus to enhance PRC2 activity, which are recruited to the specific chromatin loci by transcription factors (TF), cis-regulatory elements (orange bar), and noncoding RNAs (blue waving lines) to deposit H3K27me3 and silence master transcription regulators controlling diverse developmental programs. This molecular mechanism orchestrates plant developmental fates, organogenesis, patterning and provides a direct and global mechanistic connection between glucose-TOR signalling and development. b, The Glucose-TOR-FIE-PRC2-FLC relay overrides the default vegetative developmental program to promote flowering. This molecular mechanism may underly the link between glucose and vernalization-mediated floral transition stimulated by prolonged cold exposure.
Supplementary information
Supplementary Table 1
ChIP-Rx–seq peaks of H3K27me3 and H3K9me2 in WT and tor-es. a, List of H3K27me3 peaks in WT and tor-es. b, List of H3K9me2 peaks in WT and tor-es. The ChIP-Rx–seq data are normalized with an exogenous reference genome. The number of reads in the table indicates reference-adjusted RPM (RRPM).
Supplementary Table 2
ChIP-Rx–seq peaks of H3K27me3 in seedlings without and with TOR inhibitor treatments. The ChIP-Rx–seq data are normalized with an exogenous reference genome. The number of reads in the table indicates reference-adjusted RPM (RRPM).
Supplementary Table 3
ChIP-Rx–seq peaks of H3K27me3 in WT, fie-amiR-es, GFP-FIE/fie and SSTS/AAAA/fie. The ChIP-Rx–seq data are normalized with an exogenous reference genome. The number of reads in the table indicates reference-adjusted RPM (RRPM).
Supplementary Table 4
Differentially expressed genes in WT, fie-amiR-es, GFP-FIE/fie and SSTS/AAAA/fie. Transcriptome data sets were generated from triplicate biological samples of plants at two weeks by RNA-seq analyses. The RNA expression data were normalized to the value in WT.
Supplementary Table 5
Target genes of TOR-FIE signalling marked by H3K27me3 and upregulated in SSTS/AAAA/fie. The RNA expression data were normalized to the value in WT.
Supplementary Table 6
Gene ontology analysis of upregulated H3K27me3 target genes in SSTS/AAAA/fie plants. Fisher’s exact test was used by the BiNGO to identify GO terms that are significantly over-represented with the compiled gene list. FDR, false discovery rate. The categories in biological process with fold enrichment > 2 and FDR< 10-10 were selected and presented.
Supplementary Table 7
Transcription factors marked by H3K27me3 and upregulated in SSTS/AAAA/fie plants.
Supplementary Table 8
Oligonucleotides used in this study.
Supplementary Video 1
Live imaging of the dynamic nuclear translocation of GFP-FIE prompted by glucose after starvation in the root elongation zone. (MOV 1,174 kb) Plants were grown in liquid sugar-free medium for 5 days before glucose stimulation. Plants for imaging were transferred into 35 mm glass bottom dish with 20 mm micro-well filled with 300 μl liquid 0.5× MS medium with 25 mM glucose and immediately used for time-lapse live imaging with 10 min intervals for 6 h.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ye, R., Wang, M., Du, H. et al. Glucose-driven TOR–FIE–PRC2 signalling controls plant development. Nature 609, 986–993 (2022). https://doi.org/10.1038/s41586-022-05171-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05171-5